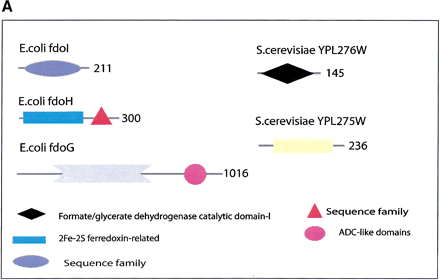
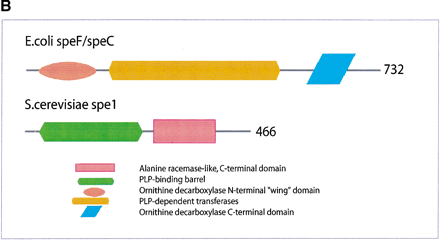
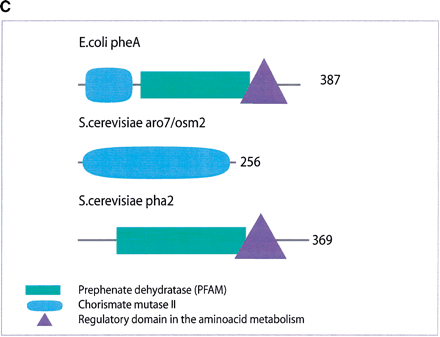
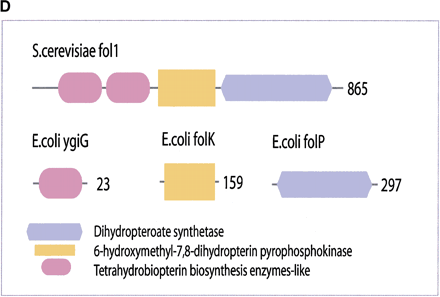
(a) This figure corresponds to the second entry in Table 8, formate dehydrogenase in Glyoxylate and dicarboxylate metabolism. This enzyme is involved in the metabolism of formate under anaerobic conditions. The reaction catalyzed is: NAD + formate → NADH + H+ + CO2. The yeast chains YPL275W and YPL276W originate from genes that are adjacent on the same yeast chromosome and make up a putative enzyme complex. The E.colichains are known to be subunits of the formate dehydrogenase complex. (b) This figure corresponds to the fifth entry in Table 6: Ornithine decarboxylase in Arginine and proline metabolism. The reaction for this enzyme is: L-ornithine CO2 + putrescine. The Escherichia coli genes speF and speC are isozymes and so share the same structure, but differ in their regulation. SpeF is the degradative form and speC is biosynthetic. (c) This figure corresponds to entry 3 in Table 7. The enzymes shown here are from the Phenylalanine, tyrosine and tryptophan biosynthesis pathway. The Escherichia coli chain, pheA, has the functions of chorismate mutase-P and prephenate dehydratase. These functions are matched by the yeast chains aro7 (chorismate mutase) and pha2 (prephenate dehydratase). The yeast chains are not known to physically interact although they are positioned consecutively in the pathway. The discrepancy in the size (a difference of 165 residues) of the chorismate mutase domain between pheA and aro7 is interesting, suggesting it either became truncated during the fusion of the yeast chains, or possibly was expanded after the fission of the E. coliprotein. The other domains involved have remained very similar in size. (d) This figure corresponds to entry 18 in Table 7. The enzymes in this example are all from folate biosynthesis. The yeast chain fol1 has the functions of dihydroneopterin aldolase, dihydro-6-hydroxymethylpterin pyrophosphokinase and dihydropteroate synthetase. YgiG is a putative kinase, folK is known as 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, and folP is 7,8-dihydropteroate synthase. Given the structural similarity between ygiG and the first two domains of fol1 it seems likely that these two are functionally equivalent, making ygiG dihydroneopterin aldolase. TheE. coli enzymes are consecutive in the pathway.











