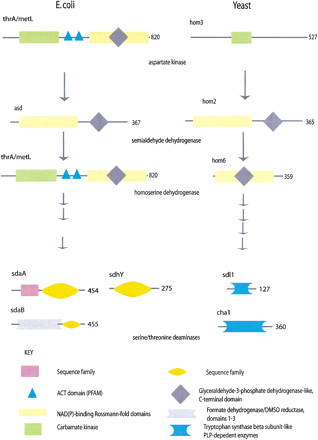
A selection of enzymes from the KEGG Glycine, serine and threonine metabolism pathway in Escherichia coli and yeast. The domain architectures of selected enzymes from this pathway are shown as cartoons along polypeptide chains represented as black lines. Domains are assigned from structure or sequence domain databases, or identified by simple pairwise sequence similarity; these latter domains are described as belonging to ‘sequence families’. Domains can be inserted into other domains such as the Glyceraldehyde-3-phosphate dehydrogenase domain into the NAD(P)-binding Rossmann fold domains in thrA and metL. These two gene products contain the domains and catalyze the reactions of both the yeast hom3 and hom6, and are thus likely to have evolved by gene fusion. Other enzymes are identical in domain architecture, such as asd and hom2. The last enzymes on the diagram, for which there are three isozymes in E. coli and two in yeast, catalyze the same reaction, but do not have any shared domains. A nonorthologous displacement has occurred among these enzymes.











