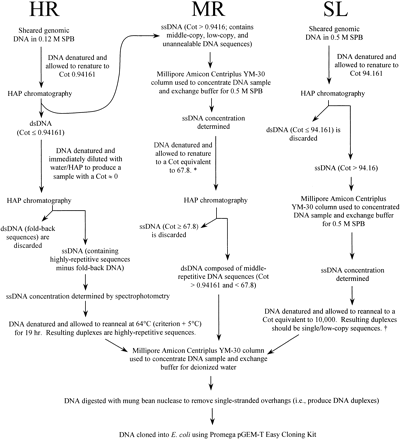
Overview of the steps involved in cloning HR, MR, and SL Cot components. DNA was denatured by heating samples in boiling water for 5–10 min. For samples in a particular sodium phosphate buffer (SPB), renaturation was allowed to occur at the criterion (Tm − 25°C) unless noted otherwise (see Britten et al. 1974 for details). Single-stranded DNA (ssDNA) and double-stranded (dsDNA) were separated using hydroxyapatite (HAP) chromatography. To attain the equivalent of a specific Cot value when starting with an isolated Cot fraction, the desired Cot value should be multiplied by the fraction of the genome remaining single-stranded at the Cot value of the starting material (see Hood et al. 1975). This principle was employed once in the isolation of the MR component (*) and once in the SL component isolation (†): * From the Cot curve, 0.67 of the genome is single-stranded at a Cot of 0.94161. To achieve renaturation equivalent to Cot 67.8 with whole genomic DNA, the Cot >0.94161 DNA was renatured to a Cot of (67.8 × 0.67 =) 45.4.† From the Cot curve, 0.28 of the genome is single-stranded at a Cot of 94.161. To achieve renaturation equivalent to Cot 10,000 with whole genomic DNA, the Cot >94.161 DNA was renatured to a Cot of (10,000 × 0.28 =) 2800.











