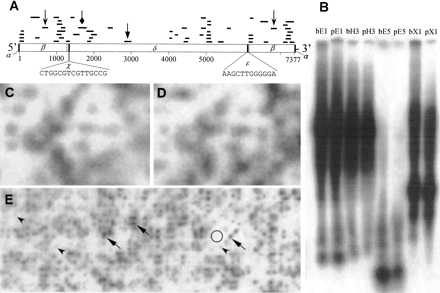
Retrosor-6. (A) The structure of Retrosor-6and the distribution of the 41 sorghum Cot clones with primary homology to Retrosor-6. Retroelement features of Retrosor-6include (α) duplicated target site sequences flanking both ends of the sequence, (β) long terminal repeats (LTRs) (bases 1–1279 and 6098–7377) with the canonical LTR start/end nucleotides 5′-TG…CA-3′, (χ) a primer binding site complementary to the plant tRNA for asparagine (bases 1286–1301), (δ) an internal sequence region with homology to ORF1 of the Arabidopsisgypsy-type retroelement Athila, and (ε) a polypurine tract (bases 6083–6095) (see Murphy et al. 1995 for review of retroelement structure). A scale showing distance in base pairs has been positioned underneath the diagram of the retroelement. For each Cot clone recognizing Retrosor-6, a thin black line has been placed above the retroelement marking the relative position(s) and length of the sequence shared by that clone and Retrosor-6. Because the LTRs have almost identical sequences (99.5% sequence identity), all of the clones with homology to one LTR have a similar/identical degree of homology to the other LTR. For these clones, lines have been positioned above both LTRs. (B) Hybridization of a Retrosor-6probe (diamond-headed arrow in A) to a Southern blot. The labels at the head of each lane indicate the source of DNA in that lane and the restriction enzyme with which the DNA was digested. Specifically, b = S. bicolor, p = S. propinquum, E1 = EcoRI, H3 = HindIII, E5 = EcoRV, and X1 = XbaI. The two species show essentially identical hybridization patterns and intensities. (C) S. bicolor grid probed with a sequence from the Retrosor-6LTR (chevron-headed arrow in A). (D) A grid identical to that in ‘C’ probed with a Retrosor-6 partial internal sequence (triangular-head arrow in A). The hybridization patterns observed for grids probed with the internalRetrosor-6 sequence are virtually identical to those produced by the LTR sequence probe for both S. bicolor (C andD) and S. propinquum (data not shown). (E) A section of the S. propinquum BAC grid probed with part of the LTR sequence of Retrosor-6. The number of copies ofRetrosor-6 in the S. propinquum and S. bicolor genomes were estimated as described in Table B (available in the online supplement to this article, www.genome.org) and the Methods section. The region on the grid used as “background” is enclosed within a circle. Examples of clones showing relatively intense hybridization signals are marked by arrows (triangular-heads) whereas clones with relatively weak but interpretable hybridization signals are marked by arrowheads (chevrons).











