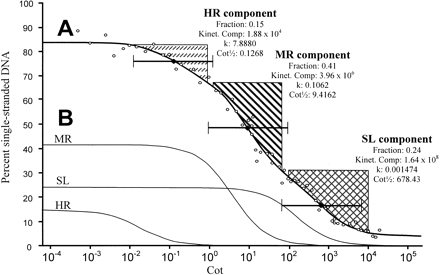
Sorghum cot analysis. (A) Complete Cot curve, data analysis, and component isolation. A least-squares curve (thick black line) was fitted through the data points (open circles) using the computer program of Pearson et al. (1977). The curve consists of highly repetitive (HR), moderately repetitive (MR), and single/low-copy (SL) components characterized by fast, intermediate, and slow reassociation, respectively. For each component, the following values have been placed to the right of the component's general location: Fraction = the proportion of the genome found in that component, Kinet. Comp. (kinetic complexity) = the length in nucleotide pairs of the longest nonrepeating sequence calculated from the Cot data, k = the observed reassociation rate in M−1•s−1, and Cot½ = the value on the abscissa of the complete Cot curve at which half the DNA in the component has reassociated. Black diamonds mark the positions on the complete Cot curve of the Cot½ values for HR, MR, and SL components. For a Cot component, 80% of the sequences in that component will renature in the “two Cot decade region” (TCDR) flanking the component's Cot½ value (brackets centered at Cot½ markers; Britten and Davidson 1985). We utilized this principle in isolating HR, MR, and SL Cot components for Cot library construction. In brief, all double-stranded DNA within a component's TCDR was isolated except for areas that overlap the TCDRs of other components (regions marked by upward vertical dashes, diagonal stripes, and crosshatching delimit areas of the curve used in HRCot, MRCot, and SLCot library construction, respectively). Note that the area isolated for use in constructing the SLCot library extends a short way past the right end of the TCDR for the SL component; this is presumably not a problem as any double-stranded DNA in the region to the right of the SL component TCDR is likely to be single-copy. (B) The predicted individual renaturation profiles of the HR component, MR component, and SL component are shown.











