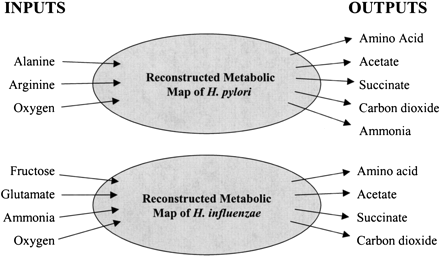
The exchange flux constraints used for the comparison of the two organisms H. influenzae and H. pylori. Note that more inputs were allowed into both systems, but the indicated inputs were the only ones used by these in silico strains. The organisms had identical output fluxes, with the exception of ammonia as a necessary output for H. pylori. The individual amino acids included in the comparison were asparagine, aspartic acid, glutamine, lysine, proline, serine, threonine, and tyrosine.











