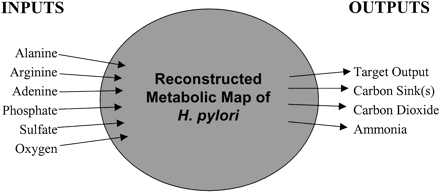
Diagrammatic representation of the in silico strain ofHelicobacter pylori and its used exchange fluxes. Other inputs from minimal medium (data not shown) were allowed, but only the inputs taken up by the system in this study are indicated. Carbon sinks include acetate, succinate, formate, and lactate. Target outputs include asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, lysine, proline, serine, threonine, tryptophan, tyrosine, the set of nonessential amino acids, and the set of nucleotides.











