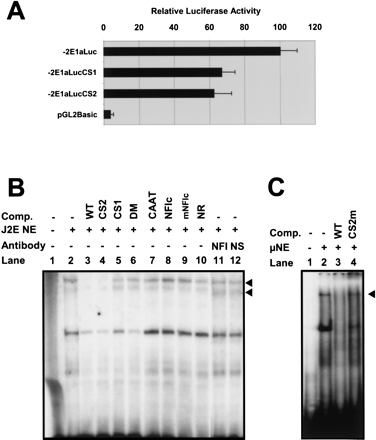
Functional analysis of the CS1 and CS2 motifs in the J2E erythroid cell line. (A) Relative luciferase activity of a wild-type mouse reporter construct containing 2 kb upstream of exon 1a (−2E1aLuc) compared with a CS1 mutant (−2E1aLucCS1), CS2 mutant (−2E1aLucCS2), and promoterless control construct (pGL2Basic). Results represent the mean of four independent experiments (a minimum of 10 luciferase values for each construct) using two different DNA preparations. (B) Gel-shift analysis identifies two specific complexes (arrowheads) binding to the CS1 motif — see text for details. (Comp) Competitor oligonucleotides; (NE) nuclear extract; (NFI) antibody to NFI; (NS) nonspecific control antibody . (C) Gel-shift analysis using a rapid protocol for nuclear extract preparation identifies a high molecular weight complex (arrowhead) binding to the CS2 site. (Comp) Competitor oligonucleotides; (NE) nuclear extract. Details of oligos used in B and C can be found in the Methods section.











