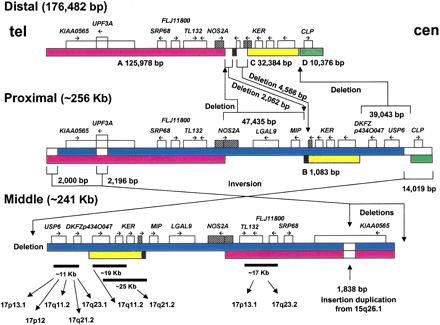
Sequence-based genomic structure of the SMS-REPs. There are four regions of sequence identity >95% between the proximal and the distal SMS-REPs (A, B, C, and D). The A (red), B (black), and C (yellow) sequence blocks have >98% identity between distal and proximal REPs; the D regions (green) show >95% identity. Blue represents the regions of homology between proximal and middle SMS-REPs. The proximal copy is the largest and is localized in the same orientations as the distal copy. The middle SMS-REP shows almost the same sequence and structure as the proximal copy except for two terminal deletions, anUPF3A gene interstitial deletion and a small (∼2 kb) insertional duplication. However, it is inverted with respect to proximal and distal SMS-REPs. SMS-REP-specific CLP,TRE, and SRP cis-morphisms (Table 1) were confirmed by DNA sequencing. Fourteen genes/pseudogenes were found and are summarized in Table 2. The two additional KER copies in distal SMS-REP represent repeated fragments of the KERpseudogenes, the accession numbers of which are given in Table 2. Crosshatched areas (NOS2A in the proximal and KER[M22927] in the distal) denote two genes spanning the high homology and nonhomology area between the distal and proximal, which suggest a three-step event for the hypothetical model of the evolution of the SMS-REPs (see text). At the bottom, the chromosome 17 distribution of fragments of SMS-REP, which constitutes a chromosome 17 low-copy repeat, LCR17, is shown. The above data were obtained throughBLAST analysis of sequence database.











