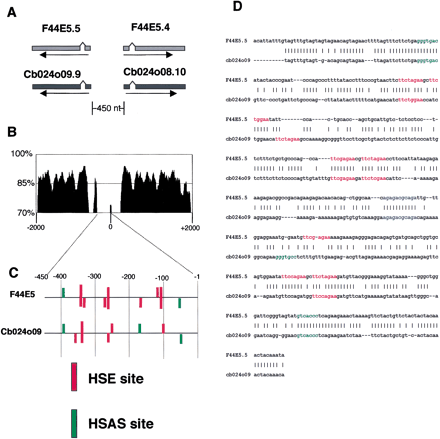
Comparison of the orthologous Caenorhabditis elegans andCaenorhabditis briggsae genes. (A) Structures of two Hsp70 protein family genes from C. elegans and corresponding orthologs from C. briggsae. The two top genes (lighter gray) are from C. elegans and the bottom two (dark gray) are from C. briggsae. (B) Sequence similarity comparison of C. elegans and C. briggsaegenes shown in A using the VISTA alignment method. The percentage of sequence conservation over every 100 nt window is indicated. Note large sequence similarity decreases in noncoding regions. (C) Heat shock element (HSE) and heat shock associated site (HSAS) DNA site positions and strengths in the promoter regions of the divergently expressed Hsp70 genes shown in A. Vertical bars represent the DNA sites and horizontal lines the DNA sequences. Height of the vertical bars are proportional to the scores calculated by the Patser program. (D) Sequence alignment of promoter regions of Hsp70 genes shown in A, using the GLASS alignment algorithm. HSEs are indicated in red and HSAS elements in green. Also highlighted in blue is a well-conserved sequence also found in thehsp-16–2/−41 promoter region (see Fig. 4A).











