Biomedical Applications and Studies of Molecular Evolution: A Proposal for a Primate Genomic Library Resource
- 1Department of Genetics, Center for Computational Genomics, and Center for Human Genetics, Case Western Reserve University School of Medicine and University Hospitals of Cleveland, Cleveland, Ohio 44106, USA; 2BACPAC Resources, Children's Hospital of Oakland Research Institute, Bruce Lyon Memorial Research Building, Oakland, California 94609, USA
Abstract
The anticipated completion of two of the most biomedically relevant genomes, mouse and human, within the next three years provides an unparalleled opportunity for the large-scale exploration of genome evolution. Targeted sequencing of genomic regions in a panel of primate species and comparison to reference genomes will provide critical insight into the nature of single-base pair variation, mechanisms of chromosomal rearrangement, patterns of selection, and species adaptation. Although not recognized as model “genetic organisms” because of their longevity and low fecundity, 30 of the ∼300 primate species are targets of biomedical research. The existence of a human reference sequence and genomic primate BAC libraries greatly facilitates the recovery of genes/genomic regions of high biological interest because of an estimated maximum neutral nucleotide sequence divergence of 25%. Primate species, therefore, may be regarded as the ideal model “genomic organisms”. Based on existing BAC library resources, we propose the construction of a panel of primate BAC libraries from phylogenetic anchor species for the purpose of comparative medicine as well as studies of genome evolution.
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: J. Rogers, C. Chiu, M. Olson, L. Williams, J. Erwin, M. Rocchio, V. Casagrande, O. Ryder, J. Allman, C. Williams, and members of the La Jolla Initiative on Human Origins.]
Importance
The study of the pattern and nature of sequence variation and its association with phenotype is central to the field of genetics. The human and mouse genomes are anticipated to be among the most completely and well-annotated genomes to be sequenced, thus providing important groundwork for future genetic and genomic studies (Collins et al. 1998). The mouse and human species are estimated to have diverged 80–100 million years ago (mya) (Kumar and Hedges 1998). Between these two species, most coding regions show a high degree of sequence similarity such that the majority of orthologous genes can readily be identified (Makalowski et al. 1996). In contrast, most noncoding regions have diverged rapidly because of the simple decay of nucleotide sequence not under selective constraint and processes of duplication, deletion, and retrotransposition. The former processes are essential to understanding the nature and pattern of single nucleotide polymorphism in contemporary human populations (Kaessmann et al. 1999;Mathews et al. 2001). Other changes (large-scale events) are important sources for sporadic and recurrent forms of genetic and genomic disease (Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002). The molecular bases and proclivity of specific regions subject to these complex genomic mutational events are not well understood. The comparison of mouse and human genomes alone will not be sufficient to understand the origin and evolution of the complex series of events (Dehal et al. 2001) that have been responsible for this variation. An assessment of genomic variation among a panel of nonhuman primate species is, therefore, required for understanding the bulk of evolutionary change between these two species and its phenotypic relevance.
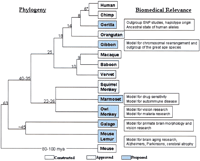
We propose the construction of a series of BAC libraries from a collection of primate species that have diverged from human at defined intervals over evolutionary time. The principal objectives will be the comparative sequencing of regions of high, biological interest (rapidly evolving genes and biomedically relevant regions) and an understanding of the pattern of genomic mutation/change over evolutionary time with respect to the human genome. Two criteria were considered in the selection of primate species: (1) its position within the species phylogeny of primates to provide a temporal view of genomic mutational change and (2) the relevance of the species to the biomedical research community (Fig. 1). Representatives of phylogenetic anchor species that are relevant to the biomedical research community were given precedent. As part of the formulation of this proposal, a large cross section of more than one dozen researchers was contacted to provide consensus on the most appropriate species for this resource.
Existing, approved, and proposed primate BAC libraries. Generally accepted phylogeny and divergence times in millions of years (*) (Goodman, 1999) and biomedical relevance of proposed libraries (shaded blue) are summarized. Existing libraries (Human (RPCI-11,13,14,15), chimpanzee (RPCI43, CHORI251), macaque (CHORI250), and baboon (RPCI-41) are available from BACPAC Resources, Children's Hospital, Oakland Research Institute (http://www.chori.org/bacpac). The construction of libraries for the orangutan, squirrel monkey, and vervet monkey (a.k.a. African green monkey) were approved (01/15/02) by the BAC Library Resource Network.
General Considerations
Nonhuman primates share closer behavioral and genetic kinship with humans than any other species. Concomitantly, among mammals, they are the most suitable biomedical models for a variety of human diseases, including models of aging, stroke, heart disease, behavior, drug sensitivity, and susceptibility to parasitic infection (Austad 1997;Rogers and VandeBerg 1998). With few exceptions, nonhuman primates are not genetic organisms in the classical sense. Low reproduction rates, long developmental periods, the exorbitant costs of handling, and the animal rights movement have curtailed research. However, despite these impediments, more than 30 species of nonhuman primates (from an estimated 300) have emerged as preferred targets for biomedical research (Austad 1997; Rogers and VandeBerg 1998). These represent a refined set of model species that have been selected because of their direct applicability to a particular disease or human condition.
The availability of a well-defined human reference sequence provides a unique opportunity for an explosion of research to investigate the genetic bases of these conditions. For example, the existence of large-insert libraries allows orthologous loci to be readily identified through hybridization experiments. The limited sequence divergence among primates (<25% at the nucleotide level for neutrally evolving genomic DNA) facilitates the rapid construction of physical comparative maps based on comparisons of large-insert end-sequence to a human reference sequence. Thus, genes and genetic markers can be recovered readily from model nonhuman primate species and used for further-refined association studies within limited primate breeding programs. A genomic-based approach involving large-insert libraries provides the most practical and cost-affordable means to interrogate the genetic bases for primate traits relevant to disease and evolution.
The paleontological record of mankind and its relatives is among the most complete (Goodman et al. 1998). More than 100 yr of anthropological research have provided independent estimates of divergence among the various superfamilies of primates. Comparative sequencing of targeted regions among a panel of nonhuman primate species provides the opportunity to directly estimate rates of single base pair change, retroposition, duplication, and deletion as a function of evolutionary time (Li 1997; Chen and Li 2001). Comparative sequencing of genomic regions among nonhuman primates (Goodman 1999;Chen and Li 2001) is essential for testing models of selection. Among immunologists, for example, comparative sequencing between human and nonhuman primates has been used to provide compelling evidence for models of balancing selection regarding genes associated with human blood group antigens (Grimsley et al. 1998; O'Huigin et al. 2000). Recently published SNP studies emphasize the value of genomic sequence from nonhuman primates to determine the ancestral and derived status of human alleles (Chen and Li 2001; Kaessmann et al. 2001). Closely related species to human (chimp, gorilla, and bonobo) are particularly valuable to eliminate ambiguity with respect to the ancestral status of a common human polymorphism. Sequences from these species provide a critical backdrop for testing the impact of genetic drift and rapid expansion on the frequency and structure of contemporary human haplotypes.
Studies of karyotype evolution require the development of a series of large-insert libraries. To date, most studies have been limited to gross cytogenetic observations, which likely mask the complexity of underlying genomic events. The number of rearrangement events has only superficially been surveyed (Yunis and Prakash 1982;Haig 1999; Muller and Wienberg 2001), and the identification of underlying genetic mechanisms, which produce such rearrangements, requires breakpoint characterization at the molecular level. Specific regions of the hominoid genome evolve much more rapidly than “generic” DNA and, therefore, require a series of primate outgroup species to resolve the complexity of these regions. Processes such as Y chromosome evolution, pericentromeric duplication, subtelomeric rearrangement, and centromere repositioning necessitate the construction of these libraries. As an example, primate genomes are frequently used to determine the timing and movement of recent segmental duplications associated with chromosomal rearrangement disorders (e.g., Velocardiofacial/DiGeorge, Prader-Willi Syndrome, Smith Magenis, etc.), pericentromeric duplications, and subtelomeric rearrangements (Kuroda-Kawaguchi et al. 2001;Mefford and Trask 2002; Samonte and Eichler 2002; Stankiewicz and Lupski 2002). These regions comprise an estimated 5%–7% of the human genome and appear to experience accelerated rates of evolutionary turnover (Bailey et al. 2001). Targeted analysis of these regions in outgroup primates has been used to reconstruct the ancestral origin of several segmental duplications and to infer the series of events that have created this duplication architecture in humans and other primates (Chiu et al. 1996; Eichler et al. 1996; Trask et al. 1998; Guy et al. 2000; Johnson et al. 2001). The construction of a BAC library is necessary to survey the structure and organization of these regions over large expanses of genomic sequence (many of the duplications or sites of rearrangement are in excess of 100 kb). Comparative sequencing will provide insight into the underlying mechanisms that have predisposed to duplication-mediated rearrangements associated with human genetic disease.
Specific Species Considerations
Four primate libraries (human, chimpanzee, macaque, and baboon) have already been constructed and are publicly available as filter sets for hybridization (BACPAC resources) (Fig. 1). An additional three libraries have been approved for construction (vervet monkey, orangutan, and squirrel monkey). With the exception of the great apes and Old World monkey species (macaque, baboon, and vervet), most other primate families are poorly represented. Therefore, large-insert BAC libraries from six primate species at critical points in the primate phylogeny are proposed. Specific details regarding biomedical relevance and usage, DNA source material, and strain selection are briefly summarized for each species. In some instances, such as the tarsiers and colobine monkeys, no species were proposed, because a biomedically relevant species could not be readily identified.
1. Gorilla (Gorilla gorilla)
Relevance: The gorilla is now recognized as an outgroup to human and chimpanzee, having diverged 1–2 mya before the separation of these sister taxa (Goodman et al. 1998; Kaessmann et al. 2001).The principal use of a gorilla BAC library would be for the purpose of comparative sequencing to determine the ancestral state of single nucleotide polymorphisms. This is particularly relevant within regions of unusual selection, i.e., HLA antigen loci in which comparative sequencing has been used to resolve the evolution of immune-related genes. Many other molecular evolution studies require a third organism to root trees that include the human and chimpanzee comparison (Kaessmann et al. 2001;Mathews et al. 2001). Finally, areas of rapid evolutionary turnover (subtelomeric, pericentromeric, Y chromosome, and large low copy repeats) cannot be adequately assessed unless a large insert library of this species becomes available.
Source: Three subspecies of gorilla are recognized: western lowland, eastern lowland, and mountain gorilla. Western lowland gorillas are the most common, least endangered, and most suitable for the purposes of BAC library construction.
2. Gibbon (Hylobates concolor)
Phylogenetic rationale: The black gibbon is one of seven representative species of the lesser apes (family Hylobatidae). It represents a phylogenetic link between the great apes and the old-world monkeys. It provides a unique view of genomic temporal change between 15 and 20 mya of species separation (human and gibbon).
Biomedical rationale: This group of organisms shows an accelerated rate of karyotype evolution, compared to other primates and most mammals (Muller et al. 1997; Muller and Wienberg 2001). Comparative studies indicate an unusually large number (n >45) of chromosomal rearrangements when compared to hominoid species. The black gibbon (Hylobates concolor) shows the largest number of such derivative rearrangement events. Unlike most hominoids, these karyotypes have been subjected to a large number of fission events. Comparative sequencing of BACs would be used to understand the molecular basis for chromosomal rearrangements, i.e., the transition region and sequences that may have predisposed to such events. Detection and sequence characterization of such large-scale rearrangements require large-insert libraries to satisfactorily traverse regions enriched in common and low copy repeats sequences. Information obtained from such studies could provide valuable insight into both germline and somatic chromosomal instability associated with chromosomal rearrangement.
Source: There are at least five different species or subspecies belonging to the genus Hylobates. Hylobates concoloris proposed, because it shows the greatest amount of karyotype variation (65 conserved linkage groups) when compared to the hominoid ancestral state (n = 23) (Burt et al. 1999). This species is commonly held in captivity.
3. Marmoset (Callithrix jacchus)
Phylogenetic rationale: This organism is a member of the New World monkeys (superfamily Ceboidea), estimated to have diverged from the anthropoid common ancestor (35–40 mya). It is an anchor species of the callitrichine clade, one of seven anciently separated New World monkey clades that diverged from each other at least 18 mya (Goodman 1999). A BAC library for the squirrel monkey, another representative of the seven ancient clades, has already been approved. Combined (squirrel monkey, marmoset, and owl monkey), these three species provide a reasonable sampling of genomic diversity among the New World monkeys.
Biomedical rationale: This species is a key organism for studies related to immunity, drug sensitivity, and brain function. Its small size, fecundity, and inexpensive handling make it one of the nonhuman primate models of choice. This species is commonly used to assess the toxicological effects of various drugs and has, on occasion, been shown to be a more appropriate model than rodents in which to test adverse drug reaction or long-term side effects (Jackh et al. 1984; Carey et al. 1992). Immunological studies have shown that the marmoset immune system is a particularly good model when compared to other primates for testing antibody specificity and recognition. Marmosets have been used to develop models of multiple sclerosis, an autoimmune disease of the central nervous system (Thart et al. 2000; Genain and Hauser 2001), as well as autoimmune colitis and thyroiditis. Cloning and comparative sequencing of gene clusters associated with drug detoxification (i.e., cytochrome P450genes) and immune response (T cell receptor, immunoglobulin genes) are essential for providing an understanding of the genetic bases for these events (Mankowski et al. 1999; Schulz et al. 2001; von Budingen et al. 2001). In addition to its primary use in immunological and toxicological studies, the marmoset has been used to develop nonhuman primate models of coronary heart disease, stroke, and reproductive disease (Charnock and Poletti 1994; Marshall et al. 2000). In the case of the latter, considerable effort has been placed on cloning, sequencing, and development of expression constructs associated with hormones and their receptors (chorionic gonadotropin, oestrogen receptor, gonadotropin-releasing hormone, hydroxysteroid dehydrogenase, and prolactin receptor) (Dalrymple and Jabbour 2000;Husen et al. 2001; Millar et al. 2001; Saunders et al. 2001).
Source: The most commonly used marmoset subspecies in research isCallithrix jacchus jacchus. The animal is not endangered. Several large colonies exist within the United States, including 235 animals at the Wisconsin Regional Primate Center and ∼80 animals located at the Southwest Regional Primate Center.
4. Owl Monkey (Aotus trivirgatus)
Phylogenetic rationale: The owl monkey is a member of the New World monkey family Cebidae, which diverged ∼22–26 mya from the squirrel monkey. Sufficient genetic distance separates these two species (estimated 7% nucleotide divergence), complicating cross-species PCR amplification. The biomedical interest in this species and its divergence justify it as a separate anchor species for BAC library construction.
Biomedical rationale: In the last 10 yr, the owl monkey (Aotous trivirgatus) has emerged as an important model for studying malaria drug and vaccine development associated with Plasmodiuminfections. Extreme variability in susceptibility to sporozoite infection has been observed among different subspecies of this monkey, and these differences have been exploited to test the efficacy of various malaria treatment regimens. Identification and characterization of a wide variety of immunological genes have been a focus of recent research in an attempt to understand the genetic basis for this susceptibility to sporozoite infections (Diaz et al. 2000; Nino-Vasquez et al. 2000; Villinger et al. 2001). In addition to its role as a model of infectious disease, this species has also been used as a model to study brain structure and morphology. The main value of the owl monkey from a neurobiological perspective is that more CNS structures have been mapped electrophysiologically in them than in any other primate except the rhesus macaque. They have also been used extensively in studies of adult cortical plasticity. Considerable research especially regarding the sensory areas of the brain has been published (Ding and Casagrande 1997). Finally, because of the unique life history of these primates (e.g., monogamy and the disparity in longevity between males and females), it is anticipated that there will be continued interest in genomic studies of these animals.
Source: Aotus trivirgatus is the most commonly used species of owl monkey in biomedical research. Small colonies are maintained in several regional primate facilities. The animals are not endangered and are easily bred in captivity.
5. Malagasy Gray Mouse Lemur (Microcebus murinus)
Phylogenetic rationale: The primate order may be divided into two major divisions: prosimians and anthropoids. Ancestral prosimians diverged ∼60 mya from the primate lineage, leading to the ancestors of New World monkeys, Old World monkeys, apes, and humans. Despite a massive extinction of prosimian species in the late Eocene (50 mya), remarkable diversity still exists (43 species are currently identified). Although several species have acquired adaptive specializations to specific ecological niches, prosimian features are generally regarded as more primitive. The prosimians occupy a unique position morphologically and phylogenetically in the primate lineage (Goodman et al. 1998). Evolutionarily, they are regarded as the outgroup of all simian species and the link to more “primitive” mammalian orders (Insectivora and Chiroptera). From the perspective of molecular evolution studies, a strong argument can be made for the construction of BAC libraries from this group. There are at least two major divisions of prosimians (galago and lemur). Microcebus murinus is biomedically representative of the latter.
Biomedical rationale: Over the last 10 yr, this species has emerged as a model for aging research. The organisms are small (50–80 g), short-lived, fecund (2–3 offspring per year), and reach sexual maturity at a young age (10 mo). Microcebus shows stereotypical signs of aging, such as susceptibility to blindness, because of lens opacity, increased frequency of tumor formation, stereotypic geriatric behavioral changes, and brain lesions similar to those associated with Alzheimer's disease (AD) (Austad 1997). Histological examination of mouse lemur brains have identified the accumulation of A beta deposits within the blood vessel walls of the cortical parenchyma, reminiscent of Alzheimer's (Gilissen et al. 1999). Several molecular studies have been initiated to recover genes associated with AD and brain aging (Bons et al. 1995; Calenda et al. 1998). Interestingly, the lifespan of mouse lemurs is dependent on the number of annual photocycles that the animal experiences. The average lifespan is five annual photocycles. If the photocycle is accelerated to 8 mo in duration, the mouse lemur still lives only five cycles on average. These observations suggest that they will become important models for other studies related to the molecular mechanisms of aging (Perret 1997). Finally, in recent years, data have emerged that suggest this species may serve as a useful model for bovine spongiform encephalopathy infection (Bons et al. 1999). Its fecundity, propinquity to humans, and its usefulness in brain aging research has led to a dramatic surge in biomedical research on this species.
Source: Large research centers with 200–300 individuals are maintained, particularly in Europe (Brunoy and Paris, France). Duke University Primate Center is an international center for research on living and fossil primates. It has the largest and most diverse collection of lemurs in the United States, which includes a small cohort of mouse lemurs.
6. Galago (Otolemur garnetti)
Phylogenetic Rationale: Galagos represent the second major division of the prosimians and are estimated to have diverged from the lemurs ∼43 mya (Martin 1990). From a phylogenetic perspective, at least two different species of prosimian should be considered for BAC library construction to control for genomic/genic idiosyncrasies in either lineage, and because the lineages leading to extant lemurs and galagos are anciently separated. Furthermore, the degree of neutral sequence divergence (>10%) and extensive karyotype variability require at least two different anchor species for this suborder of primates.
Biomedical rationale: Bush babies are prosimian primates; as such, they occupy a unique position in primate evolution, because it has been argued that ancestral prosimians gave rise to both New World and Old World simians (Martin 1990). Furthermore, contemporary prosimians are believed to possess more-primitive morphological and developmental characteristics. From this perspective, the bush baby visual system represents a more basic plan from which specializations in other primate lines arose. The organization of the visual system is well segregated (Yamada et al. 1998). Its brain is small, lissencephalic, and easily studied on histological examination.
Source: Bush babies are small primates that breed well in captivity; small colonies have been established in the United States (e.g., Vanderbilt University). They are easier and cheaper to house and maintain than their larger cousins. They are not endangered.
Summary
The primary use of nonhuman primate BAC libraries would be for comparative sequencing and mapping analyses of targeted genomic regions. It is anticipated that select regions of high biological/biomedical interest (immunological genes, genes under positive Darwinian selection, regions of rapid genomic rearrangement, haplotype characterization, etc.) would be primary targets. Because of the relatively high degree of neutral genomic sequence identity between human and our closest relatives (1.5%–25%), it is unlikely that many of these libraries would be used for a complete genomic sequencing effort. The availability of a high-quality human reference sequence, however, in combination with high-quality genomic BAC libraries, is essential for cross-genome comparisons (BAC end-sequencing or fingerprint overlay against human reference sequence) to identify regions of hypervariability or regions containing genes of biomedical interest. It should be noted that a whole-genome shotgun approach (whose inserts are relatively limited in size, i.e., <10 kb) would be ineffective in the resolution of complex genomic regions as outlined above. Furthermore, despite the low level of sequence divergence (<25%), sufficient variation exists within “generic” DNA to thwart the development of contiguous sequence over large genomic DNA using PCR methodology. For example, an attempt to analyze eight regions from the orangutan X chromosome by long-range PCR amplification (amplicons ranging from 5–15 kb in length) showed 15% failure after two rounds of oligonucleotide design and amplification, based on human reference sequence (Eichler, unpubl.). Similarly, attempts to use cross-species PCR of smaller microsatellite marker amplicons have been problematic when sequence divergence exceeds ∼6% (>25 mya of separation) (Rogers et al. 2000). Consequently, anchor species from model organisms are required to facilitate both genetic and genomic studies in various branches of the primate order.
WEB SITE REFERENCES
http://www.chori.org/bacpac; BACPAC resources web site.
Acknowledgments
During the formulation of this proposal, more than one dozen primatologists, biomedical researchers, and evolutionary geneticists were contacted, including Jeffrey Rogers (SouthWest Foundation of Biomedical Research), Chi-hua Chiu (Rutgers University), Maynard Olson (University of Washington), Larry Williams (University of Alabama), Joe Erwin (Bioqual Inc.), Mariano Rocchi (University of Bari), Vivien Casagrande (Vanderbilt University), Oliver Ryder (Center for Reproduction of Endangered Species), John Allman (Caltech University), Cathy Williams (Duke University Primate Center), and a variety of members from the La Jolla Initiative on Human Origins. We thank them for their input in species selection choices and for their critical comments in the writing of this manuscript.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL eee{at}po.cwru.edu; FAX (216) 368-3432.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.250102.
- Cold Spring Harbor Laboratory Press