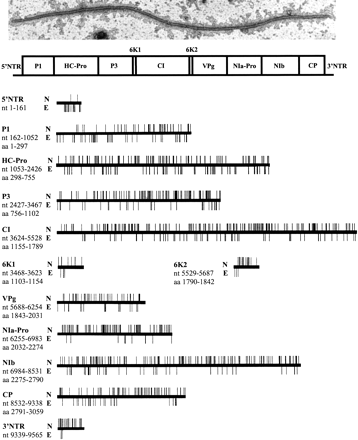
Functional map of the PVA genome. A typical PVA virion visualized by ISEM and a schematic map of the PVA polyprotein are shown at the top. For functions of the various genomic regions, see Table 1. The genomic sites nonessential (N) and essential (E) for viral genome replication and encapsidation are indicated by the bars protruding upwards and downwards, respectively, from the horizontal lines depicting the various genomic regions of PVA. The positions of the genomic regions are indicated by their first and last nucleotide (nt) or amino acid (aa). The oligonucleotides 61475 and 4141 are complementary to the 5′-proximal nucleotides of the 5′-NTR and the 3′-proximal nucleotides of the 3′-NTR, respectively, of the PVA genome, and insertions within these genomic regions could not be detected.











