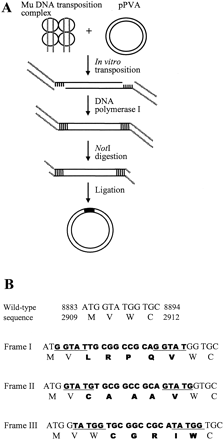
Generation of 15-bp insertion library. (A) A tetramer of MuA transposase (small circles) assembles a DNA transposition complex with a pair of modified (containing NotI site) Mu genome R end-specific DNA segments (dashed line). The complex next executes the transposition reaction by which the Mu end segments are attached to the target plasmid molecules at variable positions, creating a staggered cut. The linear two-ended transposition reaction products are isolated, and gapped reaction products are then processed with DNA polymerase I, digested with NotI, and self-ligated to produce a pool of plasmids with a 15-bp insertion. During the process, 5 bp of the target site is duplicated. (B) The amino acid sequence encoded by the 15-bp insertion (bold type) varies depending on the insertion reading frame. No wild-type amino acids are mutated or deleted and no stop codon is formed. An example of insertions into the coat protein-coding region of PVA is shown. Genomic positions are indicated both by nucleotides and amino acids (numbers). Duplicated target sites are underlined.











