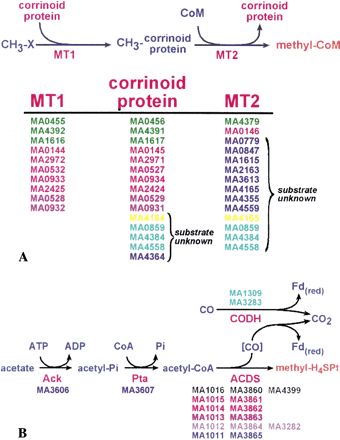
Redundant and novel genes involved in methylotrophic and acetoclastic methanogenesis. (A) The pathway for conversion of one-carbon (C-1) compounds to the central-pathway intermediate methyl-CoM (see Fig. 1). Multiple copies of substrate-specific methyltransferases (MT1 and MT2) and methylotrophic corrinoid proteins were identified for each substrate (CH3-X). The gene IDs (MA numbers) for proteins involved in the metabolism of methanol (green) and methylamine (pink); dimethylamine (red) and trimethylamine (dark red) are indicated. Five corrinoid and 12 MT2 homologs without known substrates were identified. Two shown in yellow form a putative operon; three shown in light blue comprise a paralogous family with fused corrinoid and MT2 domains. Additional MT1 homologs were not identified as these proteins do not constitute a homologous family. (B) The pathway for conversion of acetate to the central-pathway intermediate methyl-H4MPT. Two nearly identical copies of genes encoding the acetyl-CoA decarbonylase/synthase (ACDS) complex were identified (homologous genes indicated by identical color, a third lone copy of cdhA[MA4399] was also identified). Two genes (cyan) encoding single-subunit, bacterial-type CO2 dehydrogenase (CODH) proteins were also identified suggesting the possibility of exogenous CO metabolism (see text for details).











