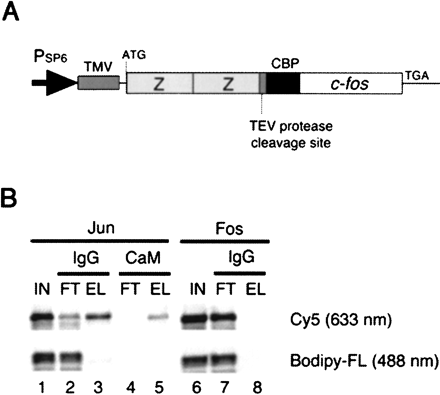
Pulldown assay. (A) A gene encoding the bait Fos (118–211 amino acids) fused with the ZZ domain and the CBP tag. TMV, tobacco mosaic virus leader sequence (Gallie et al. 1988). (B) Tandem affinity purification. The in vitro-translated Jun (lanes1–5) and Fos (lanes 6–8) proteins labeled with Cy5 (excited at 633 nm) at the carboxyl terminus and those with Bodipy-FL (488 nm) at the internal lysine residues were mixed with the bait Fos (lanes 1–6; IN, input) and captured on IgG beads (lanes 2 and 7; FT, flowthrough), washed, and eluted by TEV protease cleavage (lanes 3 and 8). The eluate was then bound to calmodulin (CaM) beads (lane 4, FT), which were washed, and eluted with EGTA (lane 5). The carboxy-terminally labeled Jun only bound to the bait Fos (Cy5, lanes3 and 5).











