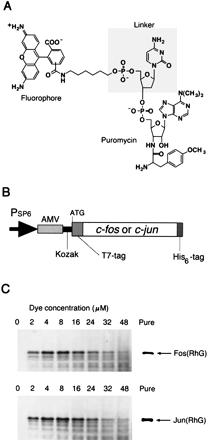
Fluorescence labeling of proteins. (A) The structure of fluorescent puromycin. A fluorophore (fluorescein, RhG, TAMRA, Cy3, or Cy5) was chemically joined to puromycin through a linker (dC, dCdC, rC, rCrC, or none). (B) Template DNA containing SP6 promoter (Morishita et al. 1999), AMV (alfalfa mosaic virus) leader sequence (Jobling and Gehrke 1987), Kozak sequence (Kozak 1986), andc-fos (118–211 amino acids) or c-jun (216–318 amino acids) gene with T7-tag (11 amino acids, MASMTGGQQMG) and His6-tag sequences. (C) RhG-labeled Fos and Jun proteins. In vitro translation reaction was carried out in the presence of the template RNA and 0–48 μM of RhG–dC–puromycin. The products were analyzed by 16.5% Tricine-SDS-PAGE with an imaging analyzer. The labeled proteins with His6-tag were purified by affinity chromatography (Pure).











