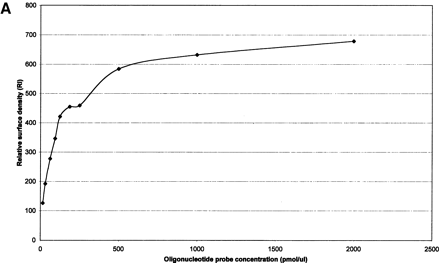
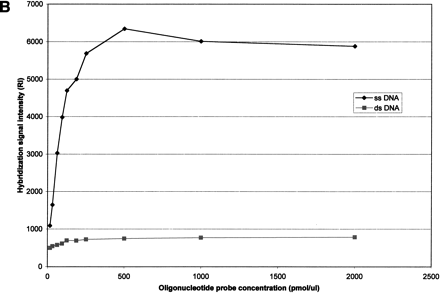
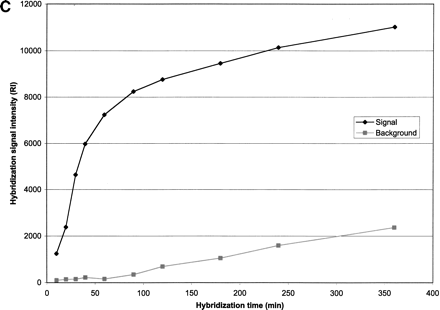
(A) Effect of oligonucleotide probe concentration on surface density of immobilized oligonucleotides. The K5 oligonucleotide probe (Table 1) was labeled with an amino group at its 5′-end and a Rhodamine dye at its 3′-end, and mixed with an oligonucleotide of the same sequence having an amino group labeled at its 5′-end (ratio of 1:10). Mixtures of oligonucleotides ranging from 15.65 to 2000 pmol/mL were spotted on activated glass slides, incubated at 37°C for 2 h and then washed with 1% NH4OH and water. (B) Effect of oligonucleotide surface density on hybridization signal intensity. A mixture of 50μL double-stranded or single-stranded Rhodamine-labeled PCR product of HLA-B exon 2 in 5×SSPE and 0.5% SDS was hybridized to the array using the K5 oligonucleotide probe (Table1) at various surface densities. (C) Hybridization kinetics. Fluorescence intensities of specific hybridization signals and nonspecific background both increase with longer hybridization time.











