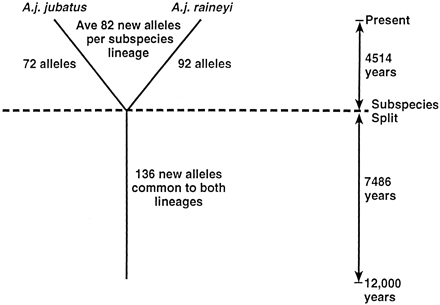
Schematic representation of new allele reconstitution at 82 microsatellite loci in cheetah populations since the homogenizing bottleneck, estimated at 12,000 yr ago (see text). Common alleles are those shared between the two geographically isolated subspecies (A. j. jubatus and A. j. raineyi). Unique alleles, which were likely produced by mutation after the geographic isolation of the subspecies, are alleles represented in one subspecies but not the other. The calculation of allele reconstitution would include a back-mutation bias, particularly as time and/or distances grow larger; however, the close agreement of the (δμ)2 estimate with the allele reconstitution calculations (see text) offers some assurance that the estimates would approximate the time period considered here.











