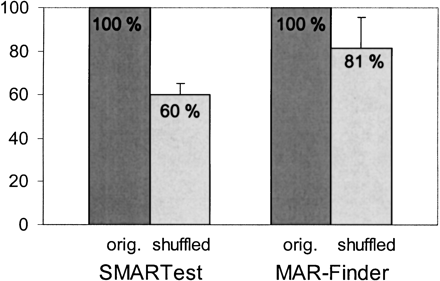
Figure 2.
Number of SMARTest and MAR-Finder predictions on five different 500 kb fragments from chromosome 22 and on shuffled sequences with a conserved local nucleotide composition. The number of predictions in the five original chromosome 22 fragments was set to 100% for both SMARTest andMAR-Finder. Mean value and standard deviation are the average of 10 experiments.











