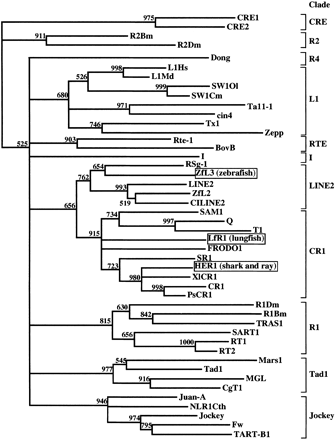
A phylogeny of families of LINEs, including LINEs that form pairs with corresponding V-SINEs. The phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei 1987), with programs in the PHYLIP package (Felsenstein 1996), using the sequences of the seven subdomains of the RTase domains of LINEs (Xiong and Eickbush 1990). Each number above a branch indicates the bootstrap value per 1000 replications and provides an indication of the statistical significance of the node. Branches with bootstrap values below 50% are reduced to polytomies. The LINEs paired with V-SINEs are boxed. Clade names (Malik et al. 1999) are shown on the right. Sources of sequences are as follows (notations refer to accession numbers unless otherwise indicated): CRE1, M33009; CRE2, U19151; Ta11–1, L47193; cin4, Y00086; Zepp, AB008896; Fw, M17214; I, M14954; Jockey, M22874; R1Dm, X51968; R2Dm, X51967; TART-B1, U14101; NLR1Cth, S59870; Q, U03849; RT1, M93690; RT2, M93691; T1, M93689; Juan-A, M95171; R1Bm, M19755; R2Bm, M16558; SART1, D85594; Dong, L08889; TRAS1, D38414; Rte-1, U00034; Tx1, M26915; L1Hs, M80340; LINE2, Repbase Update 1997 on World Wide Web URL:http://www.girinst.org/ ∼server/repbase.html; L1Md, M13002; LfR1, this study; SW1Ol, AF055642; SW1Cm, AF055643; ZfL2, Ogiwara et al. unpublished; ZfL3, AA497301 and AA497226 and this study; CiLINE2, Ogiwara et al. unpublished; RSg-1, Ogiwara et al. unpublished; HER,Ogiwara et al. 1999; SR1, U66331; CR1, U88211; PsCR1, AB005891; SAM1,U13643; FRODO1, Z70755; BovB, Z25531; XlCR1, AF027962; MGL, AF018033; Mars1, X99081; CgT1, L76169; and Tad1, L25662.











