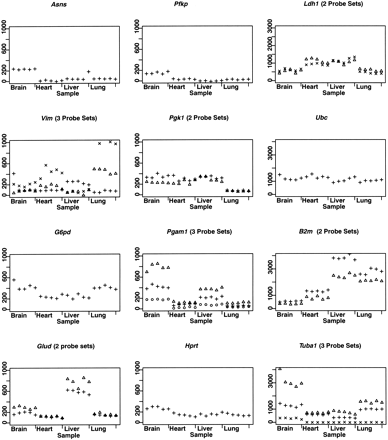
Replicate samples from four mouse tissues. RNA was extracted from the liver, heart, lung, and brain of three adult male C57BL/6 mice. To assess technical variability, we divided the RNA from each tissue of one mouse and hybridized it in replicate to three separate arrays. To assess biological variability, we hybridized RNA from identical tissues of three individual mice to three separate arrays. Points are arranged in the following order for each tissue: mouse1-replicate1, mouse1-replicate2, mouse1-replicate3, mouse2-replicate1, mouse3-replicate1. Multiple probe sets, present for Glud,Pgk1, Pgam1, and Ldh1, show consistency in measurements of expression levels across tissues. Other probe sets forTuba1, Vim, and B2m show a higher degree of variability, indicating issues inherent in probe design. Samples were normalized by mean intensity per scan.











