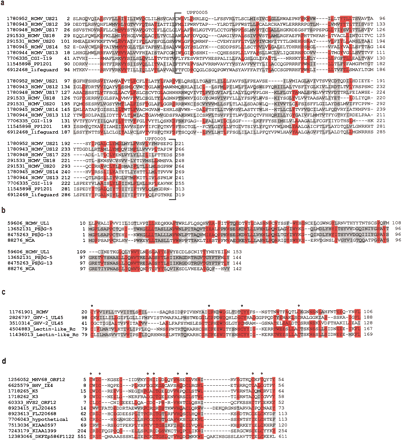
Alignment of new herpesvirus/human homologs. Proteins are labeled with GenBank identification number (GI) and a short description. Amino acids that are shaded red share identity across ≥50% of the alignment; amino acids shaded grey share similarity across ≥50% of the alignment. (a) Herpesvirus US12 protein family members, human lifeguard protein, and two additional human proteins. The Pfam UPF0005 domain is indicated. (b) HCMV UL1, two PSG proteins (PSβG 5 and 13), and one member of the carcinoembryonic antigen subfamily (NCA, nonreacting antigen). (c) A representative from each of the herpesvirus protein families found to contain C-type lectin domains and two natural killer receptors (NKG2-A). The four conserved cysteines, important for disulphide bond formation in the carbohydrate recognition domain, are indicated. (d) K3/K5 herpesvirus protein family with six human homologs. Cysteine/histidine conserved residues in the BKS (BHV-4 [bovine herpesvirus 4], KSHV, andswinepox) motif are indicated.











