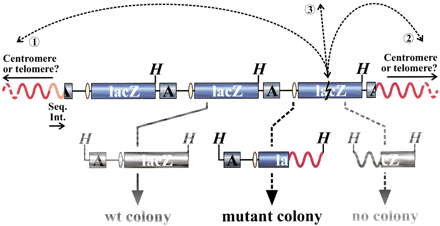
Recovery of integrated LacZ-plasmids. Each integration site comprises a concatamer of ∼10 head-to-tail organized plasmid copies (only three plasmid copies are depicted). Individual plasmid copies are released from mouse genomic DNA (red curving lines) by HindIII digestion (H indicates HindIII site). After purification, self-ligation, and transformation into Escherichia coli host cells, the plasmids are recovered in the form of ampicillin-resistant colonies. The dashed arrows represent the occurrence and detection of a hypothetical genome rearrangement with one breakpoint in a lacZ-reporter gene (lightning-sign) and one in the mouse genome 5′ (➁) or 3′ (➀) of the integration site or on another chromosome (③). When not deleted, only the upstream truncated plasmid sequence will result in a mutant colony afterHindIII digestion, as the origin of replication (yellow ellipse) and the ampicillin-resistance gene (A) are needed for selective propagation of the transformed E. coli host. The orientation of the cloned 5′ sequences of the integration sites (Seq. Int.; orange curve), indicated by the arrow, corresponds with the direction of the blue arrowheads in Fig. 3 and is arbitrary within this scheme. The physical location and orientation of the cloned 5′ integration site sequence and the recovered mouse genomic sequence in the mutant plasmid allow for characterization of the recovered genome rearrangement. See text for more details.











