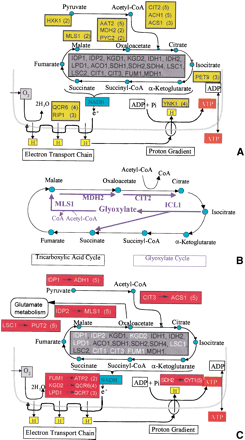
(A) Schematics of the TCA cycle, along with the oxidative phosphorilation and the ATP synthesis modules. The genes cataloged in MIPS as the TCA cycle genes for which we had expression data are listed in the central grey square. The green squares contain the FP genes that appeared in at least two out of the five cross-validation trials. (The number between parentheses next to a gene indicates the precise number of times that gene appeared as a FP.) The green squares appear next to the metabolite that the corresponding genes interact with. (B) Schematics of the glyoxylate cycle (blue arrows) that is intertwined with the TCA cycle. Of the four genes that are specific of the glyoxylate cycle, three of them appeared as FP in the TCA cycle. (C) Similar as in A, but here the red squares contain the FN (left of the arrow, also in white in the central grey square) and an arrow pointing to the gene with an expression profile that was closest to the FN. Only FNs that appeared in two or more of the cross-validation trials appear (the precise number appears in parentheses). The red squares are close to the metabolites that the enzyme associated with the FN interacts with.











