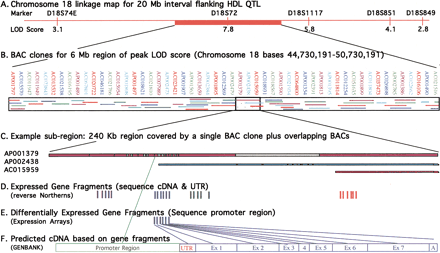
Experimental design for prioritization of genes within a linkage interval. (A) Chromosome 18 linkage map of the 20-Mb interval in the human genome flanking the equivalent position of the indicated HDL1-C QTL in the baboon genome. The top labels show the microsatellite markers, and the bottom labels indicate the multipoint LOD score for each equivalent marker in the baboon genome rounded to the nearest integer. (B) Human BAC clones covering a 6-Mb portion of the 20-Mb region of interest. Designations of the clones are shown on the top line, and coverage by each clone is shown within the boxed area. Association between clone position and designation is indicated by colors of clone names corresponding to colors of clone fragments. (C) A sample 240-Kb subregion (47,885,905–48,125,905) covered by a single BAC clone plus additional overlapping clones. Colored regions indicate complete DNA sequence data; gray regions indicate draft sequence data. Only the 5′ portions of BAC clones AP002438 and AC015959 are shown (i.e., the portions that overlap AP001379). (D) Expressed gene fragments for this region are denoted by solid vertical lines as determined by reverse Northern blot analysis. Fragments are grouped into predicted cDNAs based on GenBank homologies. Expressed genes showing no differential expression will be sequenced in cDNA and untranslated regions (UTRs) for the panel of sib-pairs. (E) For expressed gene fragments showing differential expression between animals of contrasting phenotypes, baboon BAC clones will be isolated using the gene fragments. The BAC clone promoter will be sequenced and the sequence data used to sequence promoters for the panel of sib-pairs. (F) cDNA and promoter region structures will be predicted based on sequence data comparison with GenBank database homologies.











