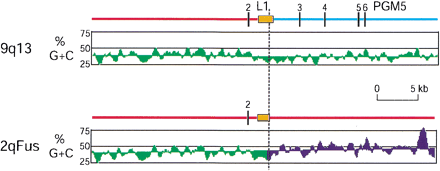
Disruption of isochore, L1 repetitive element, and PGM5 gene by the breakpoint of duplication between 9q13 and 2qFus. (Green) L isochore; (blue), H1–2 isochore. The regions shown correspond to nucleotides 381651 to 423067 in the 2qFus contig and, for 9q13, from nt 139050 in RP11–561023 to nt 9713 in RP11–88I18. The breakpoint of homology (dotted line) was determined by cross_match (http://www.genome.washington.edu/phrap_documentation.html) analysis, the L1PBa repeat was identified by RepeatMasker, and the GC-content and isochore classification were determined by GESTALT. The vertical bars are exons of the PGM5 gene (see Fan et al., 2002 for more details).











