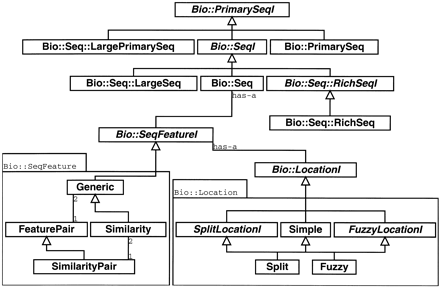
This figure shows a portion of the Bioperl object model including the interfaces (shown in italicized type) for sequences (PrimarySeqI, SeqI, RichSeqI) and their implementations PrimarySeq (general sequence), Seq (sequence with features), RichSeq (sequence with features and rich annotation), LargePrimarySeq (for sequences too large to be held in a program's memory), and LargeSeq (large sequences with features). Also included in the diagram is the sequence feature interface (SeqFeatureI) and its implementations Similarity (manage similarity information), FeaturePair (paired feature information), and SimilarityPair (paired similarity information such as a pair-wise alignment information). Additionally, the diagram shows the location objects that manage Simple (start, end, and strand information), Split (multiple start and end spots on a sequence such as a set of exons), and so-called Fuzzy locations (where start, end or span is not exact) for sequence features.











