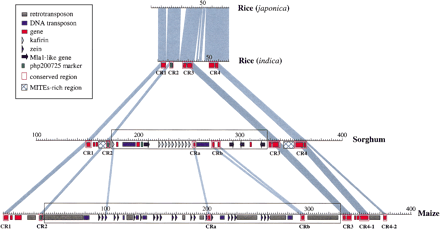
Sequence comparison of orthologous regions from rice, sorghum, and maize. Sequences of genomic regions from rice (subspeciesjaponica and indica), sorghum, and maize have been vertically arranged with a size ruler (in kilobases) for each sequence. Only orthologous regions (CR1–CR4) are highlighted. Light-blue areas indicate the homologous regions. The boxed sections between CR2 and CR3 in sorghum and maize represent an insertion relative to rice. Pink boxes along the rulers indicate conserved sequence regions across species (CR1–CR4, CRa, CRb). In maize, CR4 was divided into CR4-1 and CR4-2, reflecting an insertion relative to sorghum and rice. In sorghum, CRb was split into two pieces because of an insertion relative to maize. Green bars indicate RFLP marker php200725 and its orthologs. The second green bar in the sorghum region marks the duplication of php200725. Small triangles indicate amplified storage-protein gene copies and their orientation (22-kD zeinin maize and 22-kD kafirin in sorghum, respectively). Violet arrows indicate Mla1-like disease-resistance genes and their orientation. All other genes are indicated by small red bars. Genes without a pink outer box indicate nonconserved genes. Gray bars indicate areas of retrotransposons, and blue bars DNA transposons. Two hatched boxes in sorghum indicate two MITE-rich regions (5′ region, <1 kb/MITE; 3′ region, <2 kb/MITE). All kafirins andzeins showed homology to each other, but only the oldest copies are indicated. A summary of predicted genes and their positions is given in Table 1 for convenience.











