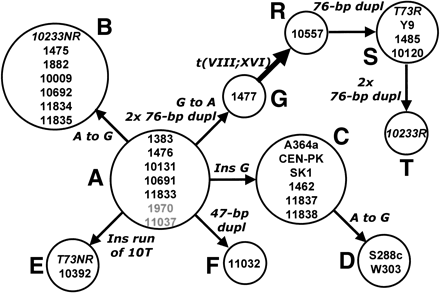
Maximum parsimony tree that minimizes the number of mutational events required to connect all the sequence variants of the ECM34gene promoter from different Saccharomyces strains N to N, nucleotide substitutions; ins, insertions; dupl; sequence duplications or repeats; 2x, a double event. A to G are nonrecombinant sequence variants, and R to T corresponds to the recombinant variants (pECM34-SSU1 or SSU1-R) generated by the unequal crossing-over involved in the translocation t(VIII;XVI). The translocation event due to unequal crossing-over between microhomology regions located in the ECM34 and SSU1 promoters is indicated by a thick arrow. Strains in gray correspond to S. pastorianus. Strains in italics are heterozygotes for the translocation and contain both a recombinant (R) and a nonrecombinant (NR) variant.











