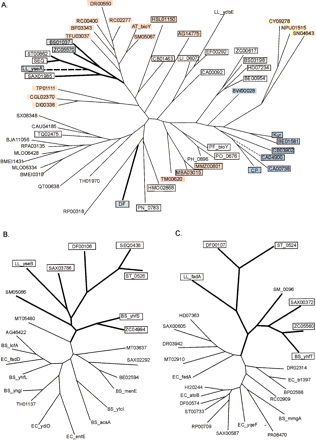
Maximum likelihood trees of the predicted biotin-related transporter BioY (A), the hypothetical long-chain-fatty acid-CoA ligase YhfT (B), and the hypothetical acetyl-CoA-acetyltransferase YhfS (C). Genes predicted to be regulated by BirA are boxed and shown in bold. The co-occurrence of the bioY,yhfS, and yhfT genes in one genome is shown by thick lines. Background colors signify: (black) single bioY gene; (blue) bioY from the biotin biosynthetic operon; (red)bioY in one operon with cbiO–cbiQ; (yellow)bioY in one operon with lspA; (magenta) bioYin the fadH–fabZ–fabK–bioY–fabD operon; (green)bioY positionally linked to the yhfS-yhfT gene pair. The bioY genes positionally linked to birA are shown by broken lines. The genome abbreviations are listed in Table 1.











