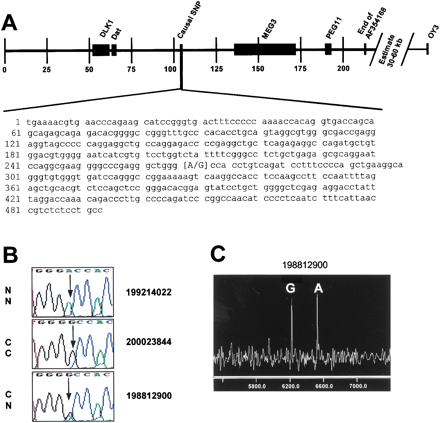
Identification of the causal base change (SNP) for CLPG. (A) Position of the identified SNP relative to previously identified candidate genes (DLK1, DAT, MEG3, and PEG11). Microsatellite marker OY3 was the previously defined telomeric boundary. Scale is set to 1 = 1000 bp of the sheep genomic sequence in GenBank accession AF354168. The SNP is at position 267 of the sheep STS sequence in accession AF401294. (B) Sequence traces representing the different genotypes present at the noted SNP. A Romanov ewe (NN at CLPG) displays the homozygous adenine residue, while the progeny-tested composite ram (CC atCLPG) displays the homozygous guanine residue. The inbred ram 198812900 (CN at CLPG) exhibits both residues at this position. (C) Results from a MALDI-TOF mass spectrometry genotyping assay for this SNP. The two alleles are represented as different mass extension products from a probe primer with the 3′ end adjacent to the SNP (Table 2). Probe extension in the presence of a specific di-deoxy termination mix creates the different analyte masses for each allele.











