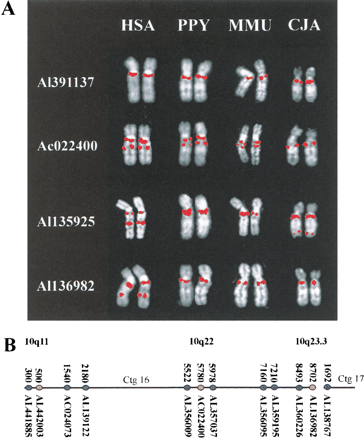
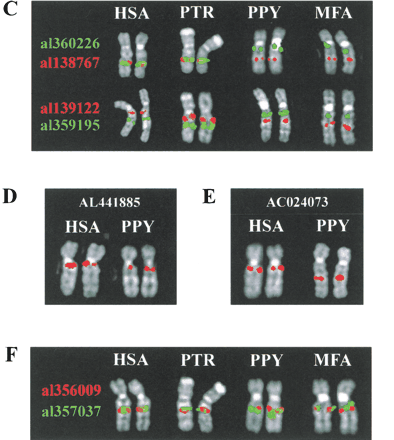
Comparative analysis of primates. Species used are as follows: hsa, human; PTR, chimpanzee; PPY, orangutan; MMU, rhesus monkey; MFA, crab-eating macaque; and CJA, common marmoset. Individual chromosomes (HSA 10 or syntenic equivalent) rather than complete metaphase spreads are shown for clarity. The species origin of each pair of chromosomes is indicated at the top of each panel. The identity of the probes used in C and F is indicated by the color of the accession number to the left of each panel. For the relative position of probes used in A, see Table 1. For the relative position of probes used in C–F, see B. (A) Comparative mapping of clones containing proximal KIAA0187paralogs. A clone containing the functional KIAA0187 gene hybridizes specifically to 10q11 in all four species. A signal is observed close to the telomere of the long arm in CJA using AL135925, indicating further lineage-specific dispersal of sequences in this clone. (B) Relative position of clones used in analyses of rearrangement. The position of each clone within Sanger Centre contigs 16 and 17 are shown in blue. The position of 10q11, 10q22, and 10q23.3 paralog clusters are shown in yellow. Contigs 16 and 17 extend from 10q11 to 10q25, with a single gap in 10q23.3. (Bentley et al. 2001). (C) Comparative analysis of clones flanking the 10q23.3 paralog cluster. (D) Delineation of proximal inversion breakpoint. (E) Comparative analysis of clones flanking the 10q22 paralog cluster.











