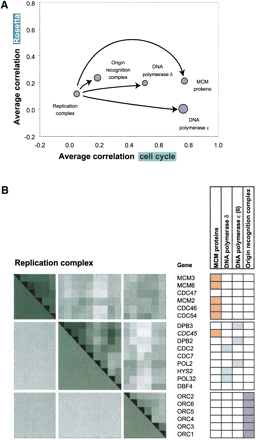
(A) A representation of the replication complex and its components on the same coordinates as the protein complexes in Figure3B. The transient replication complex can be decomposed into smaller complexes: the origin recognition complex, the MCM proteins, and the DNA polymerases δ and ɛ. Whereas the whole replication complex exhibits an average correlation close to zero (in both the cell cycle and the Rosetta data), the four smaller complexes show greater correlations in the cell cycle experiment. The four subcomplexes behave more like permanent complexes than the replication complex as a whole.
(B) The correlation coefficient matrix for the subunits of the replication complex derived from the cell cycle data. The upper triangle of the correlation matrix shows the individual correlation coefficients for particular gene pairs (with darker colors indicating higher correlations). The lower triangle shows the average correlations for subgroups of proteins (representing the MCM proteins, the two DNA polymerases, and the origin of the replication complex) within the complex as a whole. The table on the right side shows which genes belong to which subgroups in different colors. The genes were ordered with unsupervised clustering (average linkage) without regard to their classification according to the three subgroups. It can be seen that this order reflects the separation according to the subgroups very well (only the proteins in the two DNA polymerase cannot be separated into two groups). An exception is the CDC45 protein that belongs to the MCM proteins but tends to cluster with the DNA polymerases.











