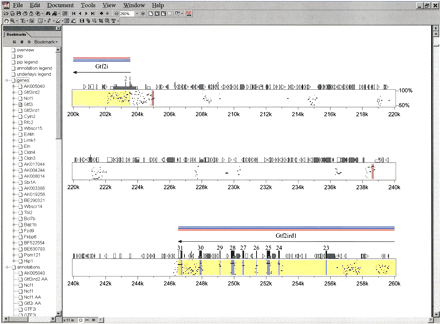
Representative portion of the percent-identity plot (PIP) comparing mouse and human sequence from the Williams syndrome (WS) region. The finished mouse sequence reported here was compared with the available orthologous human sequence using PipMaker. The complete PIP and details about the various annotations it contains are available at http://bio.cse.psu.edu/publications/desilva. Shown here is a ∼60-kb region containing portions of theGtf2i/GTF2I and Gtf2ird1/GTF2IRD1genes and the interval residing between them. Note that only gap-free segments that are ≥50% identical between mouse and human are plotted. The first two exons and last nine exons ofGtf2i/GTF2I and Gtf2ird1/GTF2IRD1, respectively, are represented by vertical rectangles and numbered accordingly; most of these exons are associated with high levels of mouse–human sequence conservation. Note the two conserved noncoding sequences at ∼205 kb and ∼239 kb (both are gap-free segments of >100 bp in length with mouse–human sequence identities of >70% and >90%, respectively, as indicated by the different colored vertical lines at those positions). Also note the various colored horizontal bars drawn above the two genes; in the actual PDF file generated byPipMaker, these bars provide direct links to relevant Internet sites (e.g., appropriate PubMed citation[s] for the gene [pink], the GenBank record containing the predicted amino acid sequence of the protein encoded by the gene [light blue], and the LocusLink entry for the gene [dark blue]). The bookmarks along the left side provide links to compiled information about the various genes and other annotations generated during the comparative analysis of these sequences.











