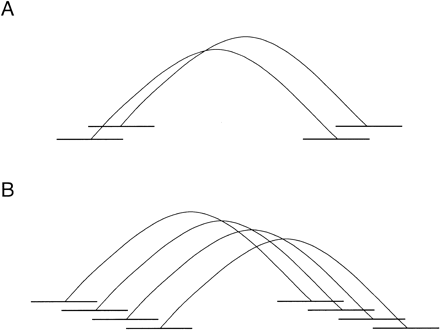
Using paired pairs of overlaps to merge reads. (A) A paired pair of overlaps. The top two reads are end sequences from one insert, and the bottom two reads are end sequences from another. The two overlaps must not imply too large a discrepancy between the insert lengths. (B) Initially, the top two pairs of reads are merged. Then the third pair of reads (from the top) is merged in, based on having an overlap with one of the top two left reads, an overlap with one of the top two right reads, and consistent insert lengths. The bottom pair is similarly merged.











