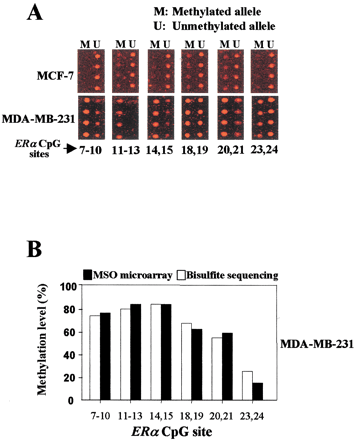
Methylation analysis of 15 CpG sites in the human ERα CpG island using MSO microarray. The nucleotide positions of these sites are shown in Fig. 2B. (A) The red fluorescence images are shown for MCF-7 and MDA-MB-231 targets. For MCF-7 images, no methylation has been detected in the interrogating sites. The same probe can differentially discriminate between M (methylated allele) and U (unmethylated allele) in the Cy5-labeled targets for MDA-MB-231 cells. Various degrees of methylation were detected in MDA-MB-231 cell lines. This experiment was independently performed three times with different target preparations. (B) Comparison of methylation levels deteteced by the methylation-specific oligonucleotide microarray with those derived from bisulfite sequencing. The histograms show the percentage of methylation levels of CpG sites analyzed in MDA-MB-231 cells. Note that the methylation levels measured by the MSO microarray showed a significant correlation with data obtained from bisulfite sequencing (P <0.001, assessed by linear regression analysis).











