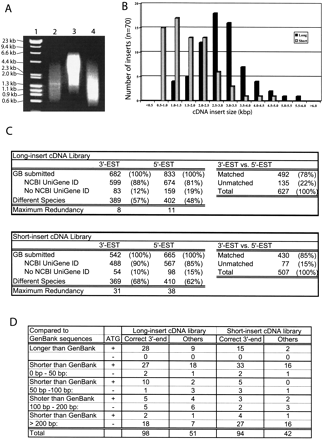
Application of the new method to the construction of newborn mouse kidney DNA libraries. (A) Size distributions of amplified cDNAs were analyzed on 0.7% agarose gels. Lane 1, DNA size marker. Lane 2, previous method. The cDNAs were tagged by LL-Sal3, and amplified by the PCR condition VI with 1 μL of LL-Sal3A primer. Lane 3, new method. The cDNAs were tagged with LL-Sal4 and amplified by the PCR condition III with 1 μL of primer S. Lane4, new method. The cDNAs were tagged with LL-Sal4 and amplified by the PCR condition V with 1 μL of primer L. (B) Comparison of size distributions of cDNA inserts between the short and long cDNA libraries. (C) Analysis of the complexity of cDNA libraries based on NCBI Mouse UniGene Index. (D) Sequence analyses of the long- and short-insert cDNA libraries. “Correct 3′-end” means that the 3′ end of cDNA sequence shows exact match to the 3′-end sequence of GenBank entry. “Others” means that the 3′ end of cDNA sequences does not match to the 3′ end sequence of GenBank entry. This can happen when either correct 3′-end sequences are not reported in the GenBank or our cDNA clones are truncated at 3′ ends. Although most cDNA clones have polyA sequence at the 3′ end, possible 3′-end truncation by Oligo(dT) mispriming cannot be excluded.











