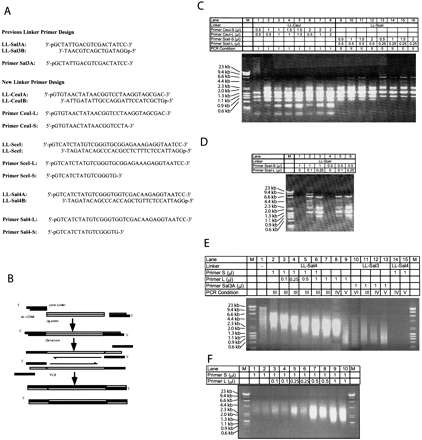
(A) Linkers and primers used in this paper. (B) Experimental procedures of lone-linker PCR. (C, D) Amplification of blunt-ended DNA size markers, λ-HindIII and φ×174-HaeIII, by various PCR conditions. Size distributions of amplified DNAs were analyzed on 0.7% agarose gels. M, DNA size marker. (E) Amplification of mouse genomic DNAs by various PCR conditions. Size distributions of amplified DNAs were analyzed on 0.7% agarose gels. M, DNA size marker; Lane 1,PvuII-digested mouse genomic DNAs (C57BL/10J); Lanes2–15, PCR products amplified from PvuII-digested mouse genomic DNA by a different combination of linkers, primers, and PCR condition. See Methods section for PCR conditions. (F) Amplification of mouse newborn kidney cDNAs by various PCR conditions. Size distributions of amplified cDNAs in duplicate were analyzed on 0.7% agarose gels.











