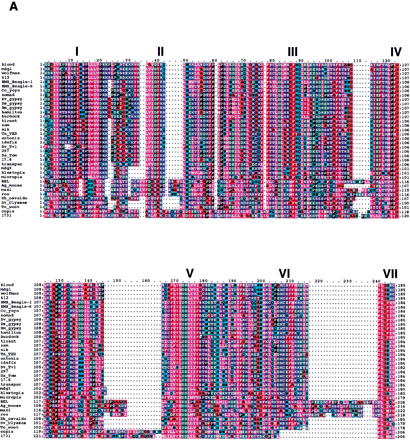
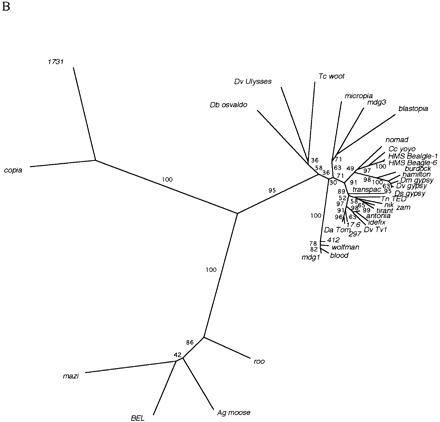
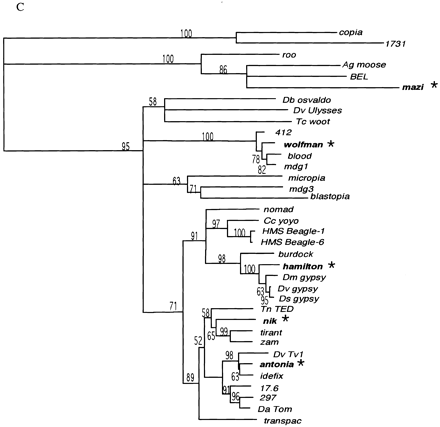
(A) Amino-acid alignment of the RT motif of Drosophila melanogaster LTR retrotransposons found in this study. The sequences were aligned using ClustalX as described in Methods. The seven conserved domains of RT (Xiong and Eickbush 1988) are indicated above the alignment. Residue coloring was performed by MacBoxshade v2.01 and is based on the similarity scheme (F, W, Y), (I, L, M, V), (P), (D, E), (G, A), (S, T, C), (N, H), (R, K), (Q). Members of a similar residue group are shaded identically. (B) Unrooted neighbor joining tree of sequences shown in A. Numbers found adjacent to branches indicate bootstrap support from 100 replicates. (C) Neighbor joining phylogenetic tree of the alignment shown in Arooted with the branch leading to the 1731 and copia lineages. Branches with bootstrap values <50 were collapsed to indicate only well-supported groups. Novel element families first characterized in this study are in boldface type and indicated by asterisks.











