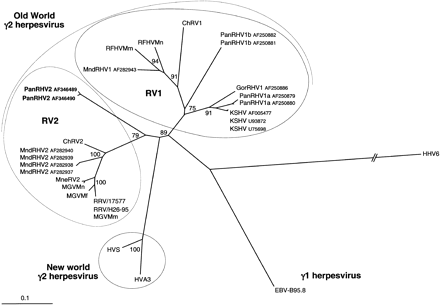
Neighbor-joining protein distance tree for the 151 amino acid residues encoded by the 454-bp fragment (primers QAHNA and GDTD1B) (Rose et al. 1997) of DNA polymerase. Sequences were aligned by usingClustalW and analyzed by using the PROTDISTand NEIGHBOR programs in PHYLIP. One hundred replica samplings were subjected to bootstrap analysis (SEQBOOT). The branch lengths are proportional to the evolutionary distance (scale bar) between the taxa. Previously published sequences included and their accession numbers are as follows: HHV6A (X83413), HVS (M31122), HVA3 (AF083424), HHV4/Epstein-Barr Virus (V01555), KSHV/HHV8 (U75698, U93872, andAF005477), PanRHV1a (AF250879 and AF250880), PanRHV1b (AF250881 andAF250882), GorRHV1 (AF250886), MndRHV1 (AF282943), MndRHV2 (AF282937–AF282940), ChRV1 (AJ251573), ChRV2 (AJ251574), RFHVMn (AF005478), RFHVMm (AF005479), RRV/17577 (AF083501), RRV/H26-95 (AF029302), MneRV2 (AF204167), Macaca γ virus strains Macaca mulatta (AF159033), Macaca fascicularis (AF159032), andMacaca nemestrina (AF159031) (named MGVMm, MGVMf, and MGVMn, respectively).











