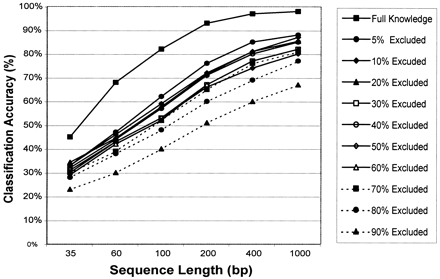
Lack-of-knowledge experiments. The genomic percentage of the genome excluded from the training phase was systematically increased and the classification accuracy was monitored. The percentage of genome excluded when training the classifier ranged from 5% to 90%. The classification accuracy in percent is represented as the arithmetic mean over all genomes and sampled sequences and is plotted on they-axis. For each genome, we sampled 100 random sequences for each sequence length, resulting in 2500 predictions for each plotted value. Different sequence lengths (35, 60, 100, 200, 400, and 100 bp) are plotted on the x-axis. Classification was based on nine-nucleotide motifs.











