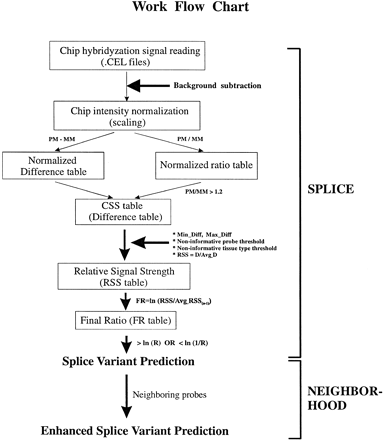
Schematic representation of the work flow chart of splice variant prediction algorithms. Raw chip hybridization intensities are extracted from Affymetrix. Cel files followed by chip background subtraction and chip intensity normalization. Normalized difference and ratio tables are generated by subtracting mismatch (MM) probe signals from perfect match (PM) probe signals and dividing PM by MM, respectively. Combined signal strength (CSS) table is created by assigning default difference value of zero to probe pairs with corresponding ratio values ≤ 1.2. To normalize expression level across tissues, relative signal strength (RSS) table is generated and followed by converting to final log ratio (FR) to further amplify the difference of relative probe signals across tissues. Candidate probes recognizing potential tissue-specific splice variants are predicted by the SPLICE algorithm. To improve the accuracy of the initial prediction, the NEIGHBORHOOD algorithm is used to assess the relative position of probes on the transcript and to generate a final prediction.











