The Evolutionary Chromosome Translocation 4;19 in Gorilla gorilla is Associated with Microduplication of the Chromosome Fragment Syntenic to Sequences Surrounding the Human Proximal CMT1A-REP
Abstract
Many genomic disorders occur as a result of chromosome rearrangements involving low-copy repeats (LCRs). To better understand the molecular basis of chromosome rearrangements, including translocations, we have investigated the mechanism of evolutionary rearrangements. In contrast to several intrachromosomal rearrangements, only two evolutionary translocations have been identified by cytogenetic analyses of humans and greater apes. Human chromosome 2 arose as a result of a telomeric fusion between acrocentric chromosomes, whereas chromosomes 4 and 19 in Gorilla gorillaare the products of a reciprocal translocation between ancestral chromosomes, syntenic to human chromosomes 5 and 17, respectively. Fluorescence in situ hybridization (FISH) was used to characterize the breakpoints of the latter translocation at the molecular level. We identified three BAC clones that span translocation breakpoints. One breakpoint occurred in the region syntenic to human chromosome 5q13.3, between the HMG-CoA reductase gene (HMGCR) and RAS p21 protein activator 1 gene (RASA1). The second breakpoint was in a region syntenic to human chromosome 17p12 containing the 24 kb region-specific low-copy repeat-proximal CMT1A-REP. Moreover, we found that the t(4;19) is associated with a submicroscopic chromosome duplication involving a 19p chromosome fragment homologous to the human chromosome region surrounding the proximal CMT1A-REP. These observations further indicate that higher order genomic architecture involving low-copy repeats resulting from genomic duplication plays a significant role in karyotypic evolution.
Several genomic disorders result from homologous recombination events between region specific low-copy repeats (LCRs). To understand mechanisms leading to chromosome rearrangements, we have investigated the basis of evolutionary breakpoints using cytogenetic approaches. The high resolution G-banding and molecular-cytogenetic comparisons of karyotypes of human (Homo sapiens, HSA), chimpanzee (Pan troglodytes, PTR andPan paniscus, PPA), gorilla (Gorilla gorilla, GGO), and orangutan (Pongo pygmaeus, PPY) showed that they differ mostly by intrachromosomal rearrangements: peri- and paracentric inversions as well as heterochromatic variations (Yunis and Prakash 1982; Jauch et al. 1992). Twelve inversions and two chromosome translocations have been identified cytogenetically, enabling distinction between human and gorilla karyotypes. After human/chimpanzee divergence, <5.5 million years ago (Mya), (Kumar and Hedges 1998), telomeric fusion of two ancestral acrocentric chromosomes resulted in the generation of human chromosome 2 (Turleau et al. 1972;Dutrilaux 1979; Yunis and Prakash 1982; Wienberg et al. 1994). The gorilla unique reciprocal translocation t(4;19) arose between ancestral chromosomes homologous to human chromosomes 5 and 17 ∼6.7 Mya (Dutrillaux et al. 1973; Yunis and Prakash 1982; Stanyon et al. 1992;Kumar and Hedges 1998). In contrast to these few rearrangements, during speciation of gibbon (Hylobatidae, HLA) at least 31, 33, and 39 chromosome translocations occurred in Hylobates concolor,Hylobates syndactylus, and Hylobates hoolock, respectively (Koehler et al. 1995a,b; Yu et al. 1997). InHylobates lar, human autosomes are divided into 51 separate HLA segments (Jauch et al. 1992).
In the majority of fluorescence in situ hybridization investigations of primate chromosomes, human chromosome painting probes have been applied (Jauch et al. 1992; Ried et al. 1993; Wienberg et al. 1997; Müller et al. 1997; Toder et al. 1998). To our knowledge, very few rearrangements in greater and lesser apes (superfamily Hominoides) have been characterized using FISH with unique sequence probes or other molecular techniques (Arnold et al. 1996; Haaf and Bray-Ward 1996; McConkey 1997;Müller et al. 2000). Recently, Nickerson and Nelson (1998) molecularly defined five evolutionary breakpoints of pericentric inversions on chimpanzee chromosomes, equivalent to human chromosomes 4, 9, and 12. Interestingly, the breakpoint on chimpanzee chromosome homologous to HSA 12q15 has been found to be associated with a submicroscopic duplication (Nickerson 2000).
The chromosome 19 breakpoint of the evolutionary gorilla t(4;19) maps to the human chromosome 17p11.2-p12 syntenic region. This human genomic region encompasses several low-copy repeat sequences and is involved in relatively frequent constitutional genomic rearrangements resulting in genomic disorders (Lupski 1998). Charcot-Marie-Tooth disease type 1A and hereditary neuropathy with liability to pressure palsy (HNPP) are caused by reciprocal duplication or deletion of a 1.4 Mb genomic fragment within 17p12, respectively, which is mediated by flanking low-copy repeats CMT1A-REPs (Reiter et al. 1996). In >90% of cases, Smith-Magenis syndrome (SMS) and dup(17)(p11.2p11.2) syndrome are caused, respectively, by reciprocal deletion and duplication of an ∼4 Mb chromosome segment in 17p11.2, also flanked by LCRs termed SMS-REPs (Chen et al. 1997; Potocki et al. 2000).
Here, we present the results of detailed molecular cytogenetic studies of the evolutionary translocation 4;19 in gorilla using FISH with human BAC, PAC, and cosmid clones. Our findings indicate that genome evolutionary breakpoints may overlap with human genomic regions susceptible to constitutional rearrangements.
RESULTS
Our molecular approach to investigate the breakpoints associated with the evolutionary reciprocal translocation yielding Gorilla gorilla chromosomes 4 and 19 was to determine which human genomic clones correspond to the syntenic chromosome regions between human and gorilla. On the basis of the comparative G-banding ideograms of human and gorilla chromosomes (Yunis and Prakash 1982), we assigned the gorilla translocation breakpoints to the human chromosome bands 5q13.3 and 17p11.2-p12. Since the 17p11.2-p12 interval contains both the 17p12 (CMT1A) and 17p11.2 (SMS) chromosomal regions, we investigated whether the evolutionary breakpoint occurred in either interval. Human genomic clones identified within these regions were then used as probes in FISH mapping studies of gorilla chromosomes (Table1). Two human clones, PAC RP1-39O20 and BAC RP11-209J20, mapping to the 17p12 (CMT1A) and 17p11.2 (SMS) regions, respectively, were co-hybridized onto gorilla chromosomes. We found that they are localized on gorilla chromosomes 4 and 19, respectively, indicating that the evolutionary breakpoint occurred between the CMT1A and SMS regions.
The Telomere–Centromere Ordered List of Human Clones Used in FISH Studies
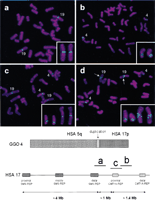
To refine the location of this important translocation breakpoint, we applied several clones mapping between these two human genomic clones as probes to gorilla chromosomes. These probes represent an ordered array of human genomic clones (arranged from the telomere to centromere direction) in proximal 17p (Table 1). Four human chromosome 17 probes (PACs RP1-140M10 and RP1-39O20, BAC RP11-726O12, and cosmid 96G3) produced one hybridization signal on gorilla chromosome 4, while four BACs (CTD-3157E16, RP11-692E18, RP11-651L9, and RP11-209J20) produced one hybridization signal on the gorilla chromosome 19 (Fig.1a,b). Two HSA 17 BACs (RP11-385D13 and RP11-640I15) showed signals on both gorilla chromosomes 4 and 19 (Fig.1c), demonstrating that they span the gorilla t(4;19) evolutionary translocation breakpoint.
Gorilla metaphase chromosomes after hybridization with human BAC clones. Below is shown gorilla chromosome 4 (GGO 4), syntenic to human chromosomes 5 and 17. The vertical arrow indicates the ∼250 kb genomic duplication, originating from the GGO 19 segment, syntenic to human chromosome, (HSA) 17p12. Smith-Magenis syndrome low-copy repeats, SMS-REPs, and Charcot-Marie-Tooth disease type 1A low copy repeats, CMT1A-REPs, in the human chromosome 17p11.2p12 region are shown (telomere to right of figure) with the relative physical position of three BAC clones used in the FISH analysis that is shownabove. (a) CTD-3157E16 (HSA 17p12), located proximal to the translocation breakpoint. (b) RP11–726O12 (HSA 17p12), located distal to the translocation breakpoint. (c) RP11-385D13 (HSA 17p12, proximal CMT1A-REP), spans the translocation breakpoint and includes the genomic duplication in gorilla. (d) CTC-428H11 (HSA 5q13.3) spans the breakpoint. Arrows within the FISH picture indicate an additional hybridization signal, which is also located on the long arm of GGO 19. This apparent duplication is located distal to the evolutionary breakpoint identified in this study. Small panels in the lower right corners demonstrate only the gorilla chromosomes 4 and 19 with positive probe hybridization signals.
Intriguingly, four nonoverlapping cosmids (4A10, 59E9, 4G3, and 123F7) (Murakami and Lupski 1996), each of which is contained entirely within two BACs spanning the translocation breakpoint, also revealed hybridization signals on both chromosomes 4 and 19, indicating that a chromosome segment of ∼250 kb, syntenic to sequences surrounding the human proximal CMT1A-REP, is duplicated on GGO 4. The human chromosome fragment, homologous to the duplicated region in gorilla, is contained within the completely sequenced BAC clones: RP11-385D13, RP11-640I15, and CTD-3157E16. Data-base searches in this human chromosome region identified several genes: HREP, NPD008/CGI-148(RP11-385D13); estrogen-responsive B box protein (ERBP), zinc finger protein (ZNF286), and ubiquitin carrier protein (E2-EPF) (RP11-640I15); Meis1-related protein 2 (MRG2), and interleukin 6 signal transducer (CTD-3157E16). Within the BAC clone RP11-385D13 we also identified another gene: CMT duplicated region transcript 1 (CDRT1), located on the telomeric side of proximal CMT1A-REP, (Fig. 2) (Inoue et al. 2001).
Schematic representation of the gorilla chromosome 19 duplicated region. Human BAC, PAC, and cosmid clones are shown below.Note that the proximal CMT1A-REP is not present in the gorilla genome. Its position in human is indicated with an arrow.
Systematic FISH mapping with several ordered BACs on human chromosome 5q13.3 (Table 2) revealed one hybridization signal on GGO 4 (BACs CTD-2156N14, CTC-431G16, RP11-414A11, and CTD-2154O16) and GGO 19 (BACs CTD-2064K14, RP11-580L9, CTC-2248H3, RP11-207B2, RP11-393G17, and RP11-639J10). BAC clone CTC-428H11 showed signals on both gorilla chromosomes 4 and 19, indicating that the breakpoint maps between HMG-CoA reductase gene (HMGCR) and RAS p21 protein activator 1 gene (RASA1) (Table 1).BLAST search results revealed that this clone also encompasses the histamine receptor H2 gene (HRH2), collagen type IV alpha-3 binding protein gene (COL4A3BP), and cyclin H gene (CCNH). Interestingly, BAC CTC-428H11 showed an additional signal, both of which are located on the long arm of GGO 4 and HSA 5, demonstrating the presence of submicroscopic duplication on these chromosomes (Fig. 1d ).
DISCUSSION
Homologous recombination between nonallelic low-copy repeated sequences has been shown to be responsible for genomic rearrangements in several different regions of the human genome (for review, seeLupski 1998; Ji et al. 2000; Shaffer and Lupski 2000). LCRs can serve as substrates for both intra- or interchromosomal exchanges, thus making the respective genomic region unstable and prone to different meiotic and apparently mitotic rearrangements (e.g., deletions, duplications, inversions, and translocations).
The human chromosome 17p11.2-p12 encompasses three Smith-Magenis syndrome low-copy repeats (SMS-REPs, ∼200 kb in size) (Chen et al. 1997) and two Charcot-Marie-Tooth disease type 1A low-copy repeats (CMT1A-REP; 24,011 bp) (Reiter et al. 1997). The proximal SMS-REP has been identified recently as the likely progenitor copy, which via duplication and duplication/inversion events at least 23.3 Mya (Kumar and Hedges 1998) resulted in distal and middle SMS-REPs, respectively (Park and Lupski 2000). Two copies of CMT1A-REP have been found in chimpanzee and human, whereas only the distal copy, which includes a portion of the COX10 gene (Reiter et al. 1997), has been identified in gorilla, indicating that it must have been duplicated after the gorilla had diverged from a common human/chimpanzee ancestor (Kiyosawa and Chance 1996; Reiter et al. 1997; Boerkoel et al. 1999;Keller et al. 1999).
Our data indicate that chromosomes 4 and 19 in gorilla, previously identified as a product of reciprocal translocation between ancestral chromosomes, apparently arose as the result of complex chromosomal rearrangement. FISH experiments demonstrated that genomic clones from human chromosomes 5q13.3 and 17p12 span the gorilla reciprocal translocation breakpoints. Genomic sequence of these human BAC clones reveals no evidence of extensive stretches of homology or low-copy repeat sequences. The results of probing with unique sequence subclones from within the BAC clones that span the breakpoint revealed hybridization signals on both gorilla chromosomes 4 and 19, indicating genome microduplication of this region.
We propose a two-step event for the origin of the t(4;19) gorilla translocation. First, a ∼250 kb chromosome segment on gorilla chromosome 19p, syntenic to the human chromosome region on 17p12 surrounding, but devoid of the proximal CMT1A-REP sequences, has been duplicated and inserted into the gorilla chromosome 4q, homologous to human chromosome 5q13.3. The duplicated and inserted fragment made the respective genomic segments unstable and susceptible to recurrent, non-allelic homologous exchanges between both chromosomes 4 and 19. Such recurrent germ line chromosome translocations in humans have been recently shown to be caused by the misaligned opposite orientated repeated satellite III sequences in the de novo Robertsonian translocations (Shaffer and Lupski 2000) and by the palindromic orientated repeated sequences in the only known recurrent non-Robertsonian constitutional translocation t(11;22) (Kurahashi et al. 2000; Edelman et al. 2001). Similar mechanism has been also proposed for the recurrent somatic translocation t(12;15)(p13;q25) in congenital fibrosarcoma (Knezevich et al. 1998; Pujana et al. 2001) and may be responsible for the origin of the pericentric inversion of chimpanzee chromosome homologous to HSA 12q15, associated with a submicroscopic duplication (Nickerson 2000). Interestingly,BLAST search results of the overlapping BACs RP11-640I15 and CTD-3157E16 revealed the presence of an ∼120 kb chromosome segment, highly homologous to the sequences flanking the proximal SMS-REP. This LCR (LCR-PSFS: ∼120-kb low copy repeat homologous to proximal SMS-REP flanking sequence) may be responsible for the origin of the described genomic duplication (Fig.2). Alternatively, the t(4;19) could have arisen only once in testis of one pre-gorilla individual and was subsequently transmitted, implanted, and accumulated as heterozygous and fixed to the homozygous state due to inbreeding in a small “bottleneck population.” The multi- versus single-exchange hypothesis could be investigated by analyzing the breakpoint polymorphisms (clusters) among three different subspecies of gorillas: Western Lowland, Eastern Lowland, and Mountain Gorilla.
We hypothesize that both chromosome events (duplication/insertion and translocation) occurred during male gametogenesis, thus increasing the number of progeny with both balanced derivative chromosomes. Supporting this, the vast majority of CMT1A duplication events in human arises during spermatogenesis (Palau et al. 1993; Lopes et al. 1997).
Recently, Mazzarella and Schlessinger (1998) estimated that up to 5%–10% of the human genome may be duplicated and Korenberg (2000)and Eichler (2000) emphasized the potential importance of chromosome duplication during primate speciation. We speculate that it was rather the described duplication and not the translocation that had an evolutionary selective advantage in homozygous or even in heterozygous carriers.
In summary, our data further indicate that higher order genomic architecture involving low-copy repeats resulting from genomic duplication plays a significant role in the genome evolution during primate speciation.
METHODS
FISH and BLAST Analyses
Because no gorilla genomic clones were commercially available, we applied human probes in FISH studies. Cosmid, PAC, and BAC probes specific for human chromosome regions 5q13.3 and 17p11.2-p12 were identified from the existing physical maps of these regions (Murakami and Lupski 1996), (http://www.ncbi.nlm.nih.gov/). BAC and PAC clones were purchased from Research Genetics and from the BACPAC Resource Center (Pieter J. de Jong, www.chori.org/bacpac). DNA was isolated from liquid cultures using PSIψClone BAC DNA Kit (Princeton Separations). The relative alignments of the selective BACs were determined byBLAST searches against the high-throughput genome sequence database (http://www.ncbi.nlm.nih.gov/blast/) and assembled using theSequencher software (Gene Codes corp.). Genes and transcripts around both breakpoints were identified by a database search of the genomic sequences using the BLASTnonredundant database (http://www.ncbi.nlm.nih.gov/blast/).
The cell line CRL 1854 of Lowland Gorilla (Gorilla gorilla) was obtained from the American Type Culture Collection (http://www.atcc.org/). FISH mapping was performed on metaphases of peripheral blood lymphocytes (human) and Epstein-Barr virus transformed lymphoblasts (gorilla) according to a modified procedure of Shaffer et al. (1997). Briefly, 1 μg of isolated BAC, PAC, or cosmid DNA was labeled by nick translation using biotin (Life Technologies-GibcoBRL) or digoxigenin (Boehringer Mannheim) labeled nucleotides. Biotin was detected with FITC-avidin (Vector Labs) and digoxigenin was detected with rhodamine-anti-digoxigenin antibodies (Sigma). Chromosomes were counterstained with DAPI diluted in Vectashield antifade (Vector Labs). Cells were viewed under a Zeiss Axioskop fluorescence microscope equipped with appropriate filter combinations. Monochromatic images were captured and pseudocolored using MacProbe 4.2.2/Power Macintosh G4 system (Perceptive Scientific Instruments).
Acknowledgments
We thank Drs. N. Boerkoel, B. Morrow, D. Nelson, and L. Shaffer for critical reviews of the manuscript. We also thank M. Withers for excellent technical assistance. S.-S.P. is supported by a fellowship from the Korean government, and K.I. by a postdoctoral fellowship from the Charcot-Marie-Tooth Association. This study was supported in part by grants from the Muscular Dystrophy Association, the National Institute of Neurological Disorders and Stroke (R01 NS27042), and the National Institute of Child Health and Human Development (POI HD39420).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL jlupski{at}bcm.tmc.edu; FAX (713) 798–5073.
-
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.181101.
-
- Received January 19, 2001.
- Accepted March 22, 2001.
- Cold Spring Harbor Laboratory Press