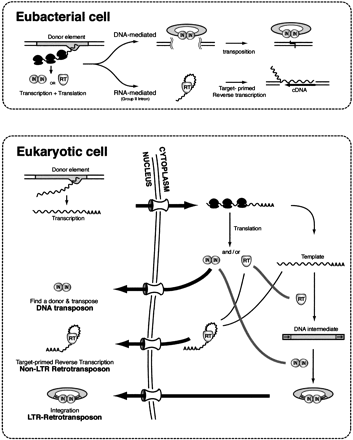
The role of the eukaryotic nucleus in the evolution of long-term repeat (LTR) retrotransposons. In eubacteria, transcription and translation are coupled; thus the encoded transposase (here labeled IN for integrase) of a DNA-mediated element can immediately bind the donor element for transposition. For simplicity, we have only shown the DNA-mediated transposition reaction as cut-and-paste, but in Eubacteria, a replicative form of transposition can also occur (Mizuuchi 1992). In the case of the RNA-mediated reaction, the reverse transcriptase (RT) can immediately bind its own transcript and initiate target-primed reverse transcription. This eubacterial precursor of the eukaryotic retrotransposons is assumed to be a mobile group II intron (Cousineau et al. 1998). The situation differs for mobile elements in eukaryotes where transcription and translation are uncoupled. Synthesis of IN in the cytoplasm means that this enzyme must enter the nucleus and find a donor for transposition. This can result in the transposition of defective copies that only retain the correct terminal repeats. In the case of the non-LTR retrotransposons, the RT must drag its RNA template back into the nucleus (cis preference) or find a new RNA template. Only the former will insure the production of active copies (Wei et al. 2001). This need to stabilize the template for entry back into the nucleus (or to wait for the breakdown of the nuclear membrane during cell division) is postulated to be the selective force that enabled the evolution of the LTR retrotransposons. LTR retrotransposons utilize both RT and IN activities. First, the RNA template is reverse transcribed into a double stranded DNA template. Second, an integrase complex shuttles this complex to a target site for integration either through the nuclear membrane or during nuclear breakdown at cell division.











