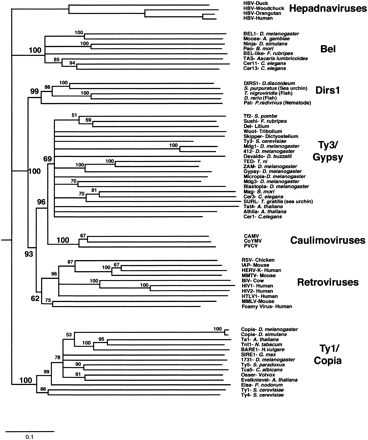
Phylogeny of the long-term repeat (LTR) retrotransposons based on their reverse transcriptase (RT) domains. The phylogram is a 50% consensus tree of the elements' RT domains (∼240 amino-acid residues) based on the neighbor-joining (NJ) method, and is rooted using non-LTR retrotransposon RTs as an outgroup (not shown). Bootstrap values are shown associated with corresponding nodes. This tree is in agreement with prior analyses (Bowen and McDonald 1999; Malik et al. 2000) except for the relative position of the Ty1/copia and hepadnaviral groups and the additional DIRS1-like sequences from sea urchin and two teleosts (accession nos. AZ181274, AL305423, and AF112374, respectively).











