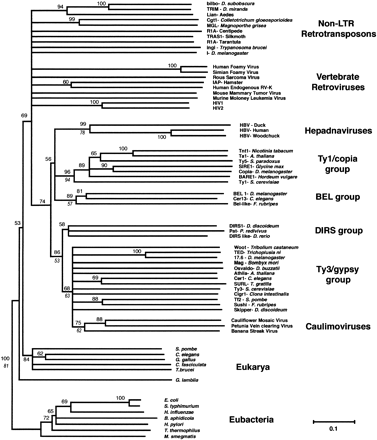
Phylogeny of the Ribonuclease HI (RNH) domains. A Neighbor-Joining (NJ) tree of the various RNH domains was performed based on the alignment of ∼140 amino acid residues in Figure 1. Bootstrap analysis was performed and nodes were collapsed to a 50% consensus. Bootstrap support (percentage from 1000 trials) for the various nodes is shown above the nodes. Maximum parsimony (MP) analysis of the RNH sequences agreed with the NJ analysis but showed lower bootstrap values for most nodes. Bootstrap values from the MP analysis for the major groupings are shown in italics if greater than 50%. The phylogeny is rooted using the eubacterial RNH domains as the outgroup. Note that the phylogenetic position of the vertebrate retroviruses is in conflict with that shown in Figure 4. All retroelement sequences are readily accessible from GenBank and previous reports (Bowen and McDonald 1999;Malik and Eickbush 1999; Malik et al. 2000).











