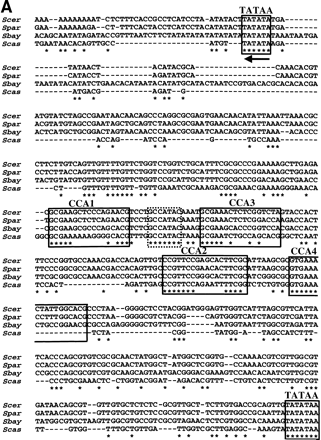
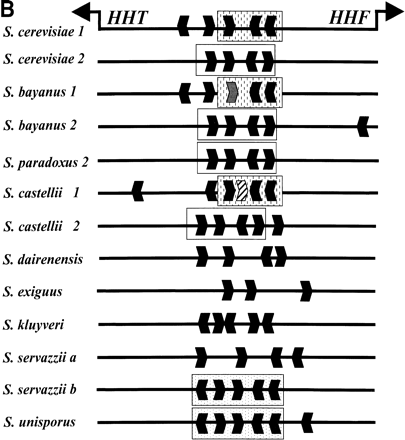

Conserved sequence elements of the HHT2–HHF2 promoter. (A) A CLUSTALW alignment of promoter sequences from S. cerevisiae, S. paradoxus, S. bayanusand S. castellii. Putative TATA boxes are boxed and underlined with arrows, indicating the direction of transcription. A conserved heptamer is enclosed in a dashed box. CCA box 1 and 2 representing elements identified in S. cerevisiae (Freeman et al. 1992) are boxed and numbered. CCA box 3 and 4 are related to the CCA box. Only part of the S. castelli CCA box 4 is captured in this alignment. (B) The location and orientation of putative CCA boxes (identified by AlignAce, Roth et al. 1998) inSaccharomyces promoter sequences. Sequences clearly orthologous to S. cerevisiae Copy 1 or Copy 2 are labeled. Unclear cases are unlabeled (if one copy was obtained from the species) or labeled as a and b (if two copies were obtained). CCA box related elements are shown as black hexagons that depict the direction of the elements. The unfilled, stippled, and vertically dashed boxes depict closely related sequences where the CCA box elements clearly align by pairwise (BESTFIT) or multiple sequence alignments (CLUSTALW). Note the extra CCA box within the S. castellii sequence (striped hexagon). The gray hexagon (in S. bayanus sequence 1) depicts an excellent CCA box that contains a single nucleotide insertion and was not identified byAlignAce, but is apparent in multiple sequence alignments. (C) A “logo” consensus for the CCA box based on 60 related sites identified by AlignAce.











