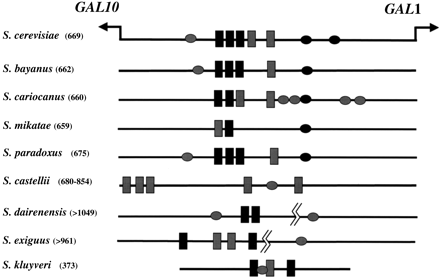
The location of Gal4 and Mig1 regulatory elements in theGAL1–GAL10 promoter sequences from many Saccharomycesspecies. The length of the promoter (where known) is shown in parentheses. Gal4 binding sites are shown as black boxes with gray boxes indicating putative binding sites that are altered by one nucleotide from the consensus site (CGGN11CCG). Potential Mig1 binding sites are shown as ellipses. Black ellipses refer to experimentally verified Mig1 sites (in S. cerevisiae) or Mig1 sites that clearly align to S. cerevisiae sites in pairwise alignments. Gray ellipses refer to putative Mig1 sites (CCCC followed by AT-rich sequence).











