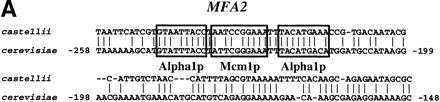
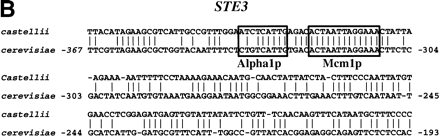

Known functional elements are conserved in BLASTNalignments between S. cerevisiae promoters and orthologous sequences of sensu lato species. (A) An alignment ofMFA2 promoter sequences from S. castellii and S. cerevisiae with known binding sites for Alpha1 and Mcm1 boxed. (B) An alignment of STE3 promoter sequences fromS. castellii and S. cerevisiae with known binding sites for Alpha1 and Mcm1 boxed. (C) An alignment of theATP14 promoter sequences from S. kluyveri and S. cerevisiae showing three conserved runs of sequence, one of which includes a consensus Hap2 (ACCAATNA) binding site. Numbers refer to the position of the S. cerevisiae sequence relative to the ATG start codon.











