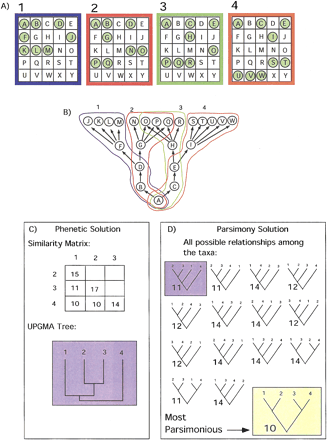
How convergence might affect tree outcome in a phenetic analysis. (A) Four arrays are shown, representing the expression profiles for four samples. Twenty-five genes, labeled A–Y, are represented by the squares in the arrays. These genes are either on (i.e., expressed) or off (i.e., not expressed). Expressed genes are shown as green dots. The hierarchy in (B) depicts a situation in which the gene expressions in the samples are related to each other in a hierarchical pattern in which two distinct expression pathways in samples 2 and 3 converge. The events that occurred in each sample to produce its array profile are outlined blue, red, green, and orange and correspond to the array above having the same color. (C) The UPGMA (Sokal and Michener 1958) solution to the array data set. The similarity matrix is calculated by counting the number of similarly expressed genes for all sample pairs, and the tree is the UPGMA clustering solution for the similarity matrix. (D) Representation of the parsimony solution. All 15 possible trees for four samples are shown. The trees are rooted using a hypothetical ancestor with none of the 25 genes expressed. The tree lengths are calculated using only informative characters, in this case genes. The tree shaded in yellow is the most parsimonious and requires the fewest changes in expression for the 25 genes shown in (A). To understand the difference between these two classification schemes, consider this example as a search for cancer-causing genes. If the expression of C, E, B, or D causes cancer, then the phenetic classification would fail to find a meaningful grouping for cancer cells. The cladistic solution shows meaningful categories. It should be noted that if O, P, or Q causes cancer, the phenetic tree would show a meaningful grouping.











